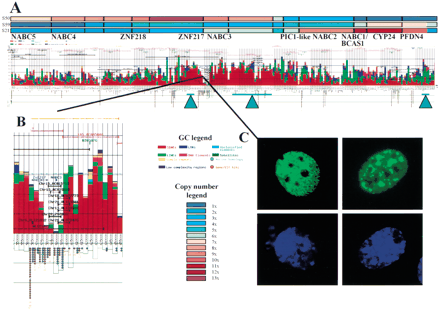
Integration of genome copy number and genome sequence information in a region of amplification at 20q13.2. (A) Genome Cryptographer (GC) analysis of a 1.2-Mb region of amplification. Average genome copy number values in selected tumors (S50, S59, S21) measured using array Comparative Genomic Hybridization (CGH) (Albertson et al. 2000) are shown as color-coded bars at the top of the figure. The array CGH data were obtained using a contig of BAC clones that now have been sequenced. Brick red lines represent public draft assemblies as of 2.1.01. Pink lines correspond to the exact size and position of the BAC clones used in the study. Densities and classification of repetitive elements are shown in color-coded cumulative bar chart above the X axis. CpG dinucleotide densities are plotted below the X axis as open green boxes. Sequence features such as genes are shown as horizontal lines above theX axis spanning the total extent of the sequence similarity. Exons are shown in bold lines. Genes and pseudogenes are represented by blue arrows pointing in the direction of transcription. The names of genes appear below the CGH copy number plot in black bold font. Total number of gene/EST hits and/or mouse identity regions are presented below the X axis as red or blue circles, respectively. Aquamarine triangles with bars, indicating the mapping resolution, mark the approximate positions of amplicon boundaries mapped by array CGH (Albertson et al. 2000), fluorescent in-situ hybridization (FISH) (Collins et al. 1998) and Southern hybridization (Collins et al., unpubl.). This figure can also be viewed athttp://shark.ucsf.edu:8080/∼stas/GR2001/index.html. (B) Enlargement of the ZNF217-NABC3 region of 20q13.2 amplification. This panel further illustrates the ability of GC to annotate features such as public draft sequence assembly (orange), BAC template locations (pink), STSs (dark green), alignment of syntenic murine sequence (light blue line), human/murine sequence identities (light blue rectangle on line), human genes (dark blue), duplications and other identities to human genomic sequence (black). The locations of genome duplications (e.g., Chr15_AC015713) are identified above the black line indicating the chromosome 20 location of each duplicon. Ratios shown beneath EST clusters correspond to the total number of EST hits/total murine EST hits. Numbers under blue circles indicate the total number of murine sequence identities per analysis interval. (C) ZNF217-EGFP fusion proteins localize to the nucleus of HeLa cells and are excluded from the nucleoli. The top two panels show localization of ZNF217-GFP fusion and the bottomtwo panels show DAPI staining of cell nuclei.











