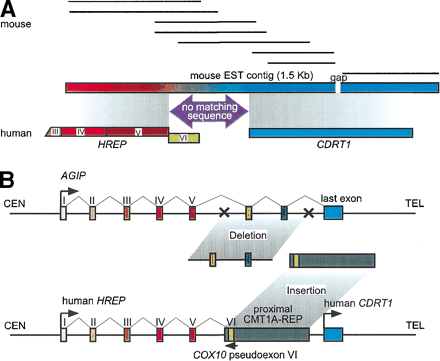
Gene evolution surrounding the proximal CMT1A–REP region. (A) A comparison of EST contigs between mouse and human. Eight mouse ESTs (shown as solid horizontal bars; GenBank accession nos. AI606691,AA089107, AA982328, AI181334, AI882388, AI429210, AI447154, andAI506955, from top to bottom) were reconstructed into a 1.5-kb contig with a 24-bp gap (horizontal rectangle with gradient colors) that aligns with two human genes, HREP (the numbers represent each exon of HREP; the exon VI does not align with the mouse EST contig) and CDRT1. Between the alignment of these two genes, there is a 269-bp region in the mouse clone that does not match any human sequence (purple arrow). The conceptual translation of this region does not identify a known protein functional motif. (B) A model for the evolution of new genes and the genomic structure surrounding the proximal CMT1A–REP region. Topfigure represents the genomic structure of a hypothetical ancient geneAGIP (Ancestral Gene before theIntegration of Proximal CMT1A–REP) modeled in mice. One or more exons originally contained in the AGIPare predicted to be lost by the integration of the proximal CMT1A–REP.Bottom figure shows human genomic structure in whichHREP and CDRT1 are separated by the inserted proximal CMT1A–REP (dark rectangle). The pseudoexon of COX10 is utilized as the last exon of HREP from the opposite direction (green box).











