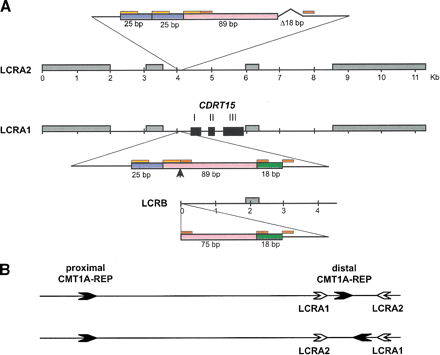
Genomic structure of LCRA1, LCRA2, and LCRB. (A) The genomic architecture of three LCRs is shown. High copy number retrotransposable elements including Alu, L1, MaLR, and MER2 type DNA elements, which are conserved between these LCR, are boxed in gray. Each exon ofCDRT15 is shown as a solid black box. A small genomic region in the 5′ upstream of CDRT15 exon I representing DNA rearrangements (enlarged) between the three LCRs. The LCRA1 has a 132-bp region that is further divided into 25 bp (blue), 89 bp (pink) and 18 bp (green) segments. There are short tandem repeat sequences, 14 bp (orange) and 9 bp (red), flanking these subdivided fragments. In comparison with the same region in the LCRA2, the 25-bp monomer is tandemly duplicated and the 18-bp fragment is deleted in LCRA2. Furthermore, the distal boundary of 4.4-kb LCRB is located in this short genomic region between 14- and 9-bp repeat (arrow). These 18 bp are present in LCRB. (B) Hypothetical inversion involving LCRA1 and LCRA2 results in flipping of distal CMT1A–REP. The proximal and distal CMT1A–REPs are depicted by thick black arrows with their relative orientation given by the directions of arrows. LCRA1 and LCRA2 are depicted as open arrows. The genome architecture as determined by sequencing this specific genomic library alone placed the CMT1A–REPs in a direct orientation and thus the region in between is susceptible to duplication/deletion. Below is shown the hypothetical orientation of CMT1A–REPs if an inversion occurs via homologous recombination using LCRA1 and LCRA2 as substrates. Note that this orientation of CMT1A–REPs will prevent the formation of duplication/deletion event (Lupski 1998b).











