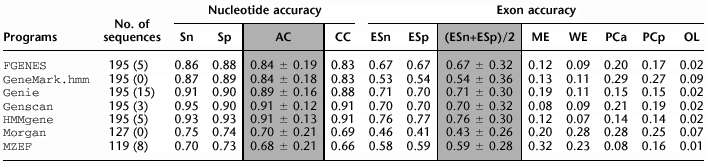
Nucleotide and Exon Level Accuracy
|
-
For each sequence in the HMR195 dataset, the exons predicted on the forward (+) strand were compared to the annotated exons. The standard measures of predictive accuracy on nucleotide and exon level were calculated for each sequence and averaged over all sequences for which they were defined. This was done separately for each of the programs tested.
-
(No. of sequences) number of sequences effectively analyzed by each program; in parentheses is the number of sequences where the absence of gene was predicted; (Sn) nucleotide level sensitivity; (Sp) nucleotide level specificity; (AC) approximate correlation; (CC) correlation coefficient; (ESn) exon level sensitivity; (ESp) exon level specificity; (ME) missed exons; (WE) wrong exons; (PCa) proportion of real exons that were partially predicted (only one exon boundary correct); (PCp) proportion of predicted exons that were only partially correct; (OL) proportion of predicted exons that overlap an actual exon. AC and (ESn+ESp)/2 are given with standard deviation.











