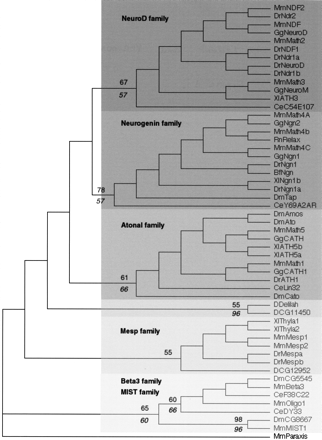
A rooted neighbor-joining (NJ) tree showing the evolutionary relationships of Atonal superfamily members. We used the closely related paraxis sequence (see Figure 2) as outgroup. The different constituting families are pointed out. Numbers above branches indicate percent support for the nodes defining the families in distance bootstrap analyses (1000 replicates). Italicized numbers below branches indicate percent reliability in a puzzle maximum-likelihood (ML) tree. The Mesp family is not supported by ML analyses. Deep nodes are not supported by resampling methods. Note that delilah andCG11450 cluster together but are not associated to any vertebrate or nematode genes. As in Figure 2, the tree shown has branch lengths that are not proportional to distance between sequences. The alignments of which this tree is based as well as maximum parsimony (MP), ML, phylogram, and bootstrapped trees can be found athttp://www.cnrs-gif.fr/cgm/evodevo/bhlh/index.html. Abbreviations are as listed in Figure 1.











