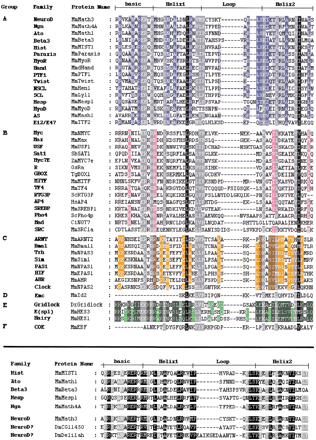
(top) Alignment of the bHLH of the 44 different families listed in Table 1 (abbreviations as in Table 1). One member per family, usually from mouse, has been selected. Designation of basic, Helix1, Loop and Helix2 follows Ferre-D'Amare et al. (1993). The different families have been grouped according to the high-order group to which they belong (Atchley and Fitch 1997; see text). The evaluation of percentage conservation within each group and through the complete multiple sequence alignment was done using the Blosum62 Similarity Scoring Table. A specific background color with three intensities is attributed to each group (dark, 100% conservation; medium, 80% or greater conserved; light, 60% or greater conserved). Dark gray and black backgrounds represent conservation through all groups. Residues with black background represent 100% conservation; residues with dark gray background represent 80% or greater conservation. (bottom) Alignment of the bHLH of the constituting members of the Atonal superfamily. One member of each family plus the two orphanDrosophila genes, CG11450 and delilah are represented. The evaluation of percentage conservation was done using the Blosum62 Similarity Scoring Table. Background intensities represent conservation (dark, 100% conservation; medium 80% or greater conserved; light, 60% or greater conserved). In this and all subsequent figures, the following abbreviations for species names are used: Bb, Branchiostoma belcheri (amphioxus); Bf,Branchiostoma floridae (amphioxus); Cc, Ceratitis capitata (a lower diptera); Ce, Caenorhabditis elegans; Ci, Ciona intestinalis (an ascidian); D and Dm,Drosophila melanogaster; Dp, Drosophila pseudoobscura; Dr, (Brachy)danio rerio (zebrafish); Ds,Drosophila simulans; Dy, Drosophila yakuba; Gg,Gallus gallus (chick); Hs, Homo sapiens; Hv,Hydra vulgaris; Jc, Juonia coenia (buckeye butterfly); Mm, Mus musculus; Ol, Oryzias latipes (Japanese medaka); Os, Oryza sativa (rice); Rn, Rattus norvegicus; Sb, Soybean (Glycina max); Sc,Saccharomyces cerevisiae (yeast); Tg, Tulipa gesneriana; Tr, Takifugu rubripes (pufferfish); Xl,Xenopus laevis; Zm, Zea mays (maize).











