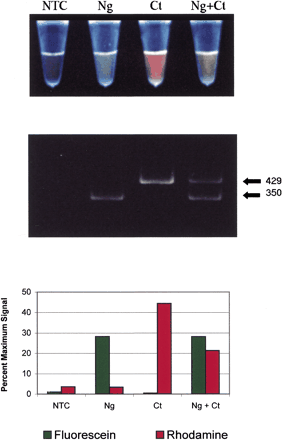
(A) Post-PCR photograph of multiplexed bacterial samples. Fromleft to right, the tubes contained no template control (NTC), 104 copies Neisseria gonorrhoeae DNA (Ng), 104 copies Chlamydia trachomatis DNA (Ct), and 104 copies of both N. gonorrhoeae DNA and C. trachomatis DNA (Ng + Ct). The tubes were illuminated from below with a 302-nm UV light, a wavelength that stimulated visible fluorescence from both dye labels. (B) The reaction products were analyzed by polyacrylamide gel electrophoresis. Two sizes of bands were visible in the UV-irradiated gel, corresponding to 429 bp and 350 bp. Although not visible in this black-and-white photograph, the 429-bp band fluoresced a bright red color and the 350-bp band was bright green. (C) The fluorescent signal of each reaction following PCR. Reaction fluorescence was normalized against the starting fluorescence of the primer in the absence of the Q-PNA.











