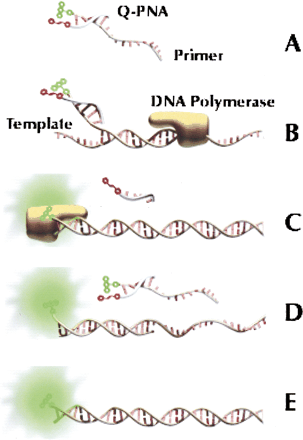
Schematic of the Q-PNA PCR principle. (A) Quencher-labeled Q-PNA hybridized to the 5′ tag sequence of a fluor-labeled forward primer to quench the fluorescence of the primer. TheTm of the Q-PNA/primer duplex is between the temperatures of the PCR annealing and extension steps. (B) In the early rounds of thermocycling, the 3′ end of the forward primer hybridizes to denatured target and DNA polymerase extends the primer. (C) The reverse primer (not shown) initiates transcription of the reverse strand, resulting in displacement of the Q-PNA and incorporation of the 13-base Q-PNA-binding region into the amplicon. (D) In subsequent annealing steps, the forward primer anneals fully to the reverse strand, reporting its presence for real-time analysis because excess primer is quenched by the Q-PNA. (E) The end product of the reaction is a doublestranded amplicon, of which one strand is fluorescently labeled.











