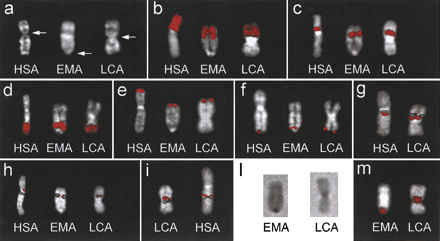
(a) Q-banded chromosome X from Homo sapiens (HSA),Eulemur macaco (EMA; black lemur), and Lemur catta(LCA, ringtailed lemur). The arrows indicate the centromere. EMA and LCA chromosomes are oriented, in all images, upside-down to match the orientation of human X. (b–f) Examples of fluorescence in situ hybridization (FISH) experiments using PCP #364 (Xp; b), PCP #377 (Xp11.2–Xp11.3; c), PCP #118 (Xq24–28;d), PCP #PGPL probe (PAR1; (e), and U130F6 cosmid clone (HSPRY3 gene, PAR2; f). (g) Cohybridization experiment using probes bA235K20 (green signal) and dJ1015G2 (red signal). Their orientation is identical in both species. (h) Probes dJ598A24 (red) and bA235K20 (green) flank the human centromere. In EMA and LCA, they appear to be almost completely overlapping. (i) Probes dJ1015G2 (red) and dJ715D6 (green) surround the LCA centromere and are well separated by the centromeric heterochromatin. In HSA, they appear much closer. (l) The C-banded chromosome X of EMA and LCA. (m) The signal of DOP-PCR amplified products from microdissected material of the EMA (left) and LCA (right) centromeric regions hybridized to EMA and LCA metaphases, respectively. Only chromosome X is shown (see text for details).











