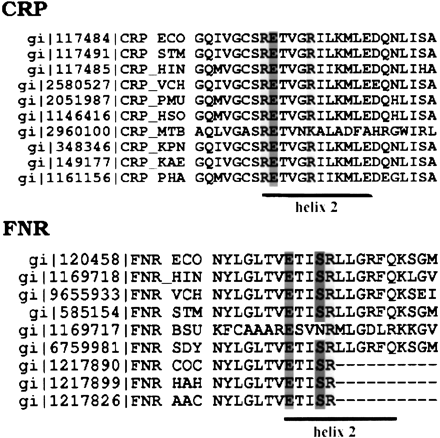
Multiple sequence alignment of CRP and FNR proteins from various bacterial genomes. Only sequences around the second helix of the helix–turn–helix motif are shown. The boundaries of the second helix are labelled with a solid line. The highly conserved RE—R motif in CRP protein and E–SR motif in FNR protein are shaded. FNR_AAC, FNR_COC, and FNR_HAH are partial sequences derived from homology cloning (Hattori et al. 1996). (AAC) Actinobacillus actinomycetemcomitans; (BSU) Bacillus subtilis; (COC)Capnocytophaga ochracea; (ECO) Escherichia coli; (HAH) Haemophilus aphrophilus; (HIN) Haemophilus influenzae; (HSO) Haemophilus somnus; (KAE)Klebsiella aerogenes; (KPN) Klebsiella pneumoniae; (MTB) Mycobacterium tuberculosis; (PHA) Pasteurella haemophilus serotype 1; (PMU) Pasteurella multocida; (SDY)Shigella dysenteriae; (STM) Salmonella typhimurium; (VCH) Vibrio cholerae. CRP, cAMP receptor protein; FNR, fumarate and nitrate reduction regulatory protein.











