Centromere on the Move
The centromeres of primate chromosomes are composed of complex arrays of alphoid sequences that are organized from tandemly repeating units of 171 bp. The ubiquity of alphoid DNA at the centromeric regions suggests a preference for repetitive DNA in the assembly of the kinetochore. Nevertheless, recent analyses of ectopic centromeres (neocentromeres) at non-alphoid-containing chromosomal sites suggest that centromeres can be “repositioned” along a chromosome through a still poorly understood epigenetic mechanism of “activation” of hitherto noncentromeric genomic DNA (Choo 1997). In this issue, Ventura et al. (2001) present evidence for centromere repositioning along the X chromosome during primate evolution that raises interesting mechanistic possibilities.
Centromere Repositioning Via Neocentromere Emergence?
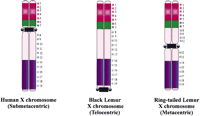
The study of Ventura et al. (2001) is based on comparison of the conservation of DNA sequences on the X chromosomes of humans and two of the Lemuridae: the black lemur and the ringtailed lemur. Lemurs are small primates related to the monkey and are found mainly in Madagascar. Unlike the submetacentric human X chromosome, the black and ringtailed lemur chromosomes are telocentric and metacentric, respectively. Ventura et al. have performed FISH analysis on the lemur X chromosomes using human chromosomal paints and a panel of BACs/cosmids spanning the entire X chromosome. Based on the relative map positions of these probes, they conclude that the lemur X chromosomes are isosequential to the human X chromosome (Fig.1). They then propose that neocentromere activation may account for the centromere repositioning that has given rise to the acrocentric, metacentric, and submetacentric configurations in the three different species (Fig. 2). Because the corresponding chromosomal regions at which the neocentromeres putatively originated do not contain the large arrays of repetitive sequences that are now found at the repositioned centromeres, Ventura et al. suggest that heterochromatic materials may have gradually accumulated at the neocentromere sites. In addition, the investigators indicate that there has been a loss of repetitive DNA materials from the original centromeres of the X chromosomes.
FISH mapping showing centromere repositioning on the X chromosomes of three different primate species. Centromere position is indicated by the black box. Colored asterisks, numbered 1–19, represent positions of the different BAC or cosmid probes used in FISH mapping analysis. Subchromosomal paints are indicated by the regions shown in red, green, or purple. Despite the different positions of the centromeres, all three chromosomes are found to be isosequential.
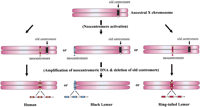
Centromere repositioning caused by activation of a neocentromere. A neocentromere may be formed de novo from a resident noncentromeric DNA on an ancestral X chromosome via epigenetic mechanisms not involving alteration to the primary nucleotide sequence of this DNA. This is then followed by the acquisition and/or amplification of repetitive DNA on the neocentromere and the inactivation and deletion of the old centromere. Alternatively, a neocentromere may be formed through the same epigenetic mechanisms following an initial transposition of a small noncentromeric genomic DNA fragment from another chromosomal site onto the ancestral X chromosome (not shown).
Ventura et al. do not favor the alternate possibility of insertional transposition of normal centromere DNA, based on the observation that no crosshybridization was detected on the human X chromosome when probes amplified from the dissected lemur centromeric segments were used in FISH analysis. However, the fact that the centromere sequences of the X chromosomes in humans and in the black lemur are common to those found on the centromeres of other chromosomes within the same cell raises the possibility that the chromosome-X centromeres may have originated by transposition or acquisition of a functional centromere from another chromosome (Fig. 3). Nevertheless, it remains possible that any transposition or acquisition of centromere repeats may have occurred subsequent to the activation of a neocentromere, therefore still identifying neocentromerization as the primary trigger for centromere repositioning (Fig. 2). The data do not allow these possibilities to be discerned.
Centromere repositioning caused by transposition of normal centromere. Centromere repositioning may occur because of transposition of a fully functional centromere, followed by loss of the old centromere on the X chromosome.
In the ringtailed lemur X chromosome, the centromere sequence is unique in regard to other centromeres, although slight homology to the interstitial heterochromatin of various chromosomes in this species, and to the pericentromeric heterochromatin of chromosomes 3 and 6 of the black lemur, was described. In this case, direct transposition of centromeric DNA is less likely to have occurred, and neocentromerization probably provides a better explanation. The low level of crosshybridization with the heterochromatin of other chromosomes suggests a possibility of transposition of noncentromeric heterochromatic DNA during the evolution of this neocentromere (Fig.2). The transposition of such DNA may have occurred either prior to the formation of a neocentromere or following an independent neocentromerization event. A recent study demonstrating that noncentromeric heterochromatic DNA is a good substrate for neocentromerization (Henikoff et al. 2000; see below) adds some support to the possibility of the transposition of heterochromatic DNA in inducing neocentromere formation. It is also possible, although unlikely, that the acquisition of such heterochromatin-related DNA may simply be an incidental occurrence that has nothing to do with the evolution of this centromere. Further discrimination between these possibilities will require the identification of the functional DNA sequences within the repositioned centromere and comparison of this sequence with those found in the corresponding euchromatic portion of the other primate X chromosomes or the heterochromatic DNA of other chromosomes.
What Triggers the Formation of a Neocentromere?
In humans, the appearance of a neocentromere and the inactivation of a pre-existing centromere go hand in hand (Choo 1997; Warburton et al. 2000). Epigenetic modifications that are known to induce higher-order chromatin repackaging have been proposed to account for the observed assembly of kinetochores on noncentromeric sequences in various species, including Drosophila and humans, suggesting that the mechanisms may be universal (Steiner and Clark 1994; du Sart et al. 1997; Williams et al. 1998). Possible epigenetic modifications include deposition of centromere DNA-binding protein that results in higher-order chromatin reorganization or occur via chemical modification of centromeric DNA or its associated histones and nonhistone proteins, such as methylation, polyribosylation, acetylation/deacetylation, phosphorylation, and ubiquitination (Choo 2000). In Drosophila, neocentromere formation has been proposed to be a consequence of cis-spreading of centromeric epigenetic modifications to juxtaposed DNA during chromosomal rearrangements (Williams et al. 1998). This cis-spreading model postulates that an altered centromere-specific chromatin conformation is being imposed onto an euchromatic region. Partridge et al. (2000) have also demonstrated the cis-spreading of fission yeast centromere proteins and suggested that the plasticity in centromere formation may be mediated in part by spreading of chromatin-associated proteins beyond the nucleation point, possibly via underacetylated or methylated chromatin. However, it is conceivable that activation may also occur via trans-spreading of epigenetic information through interaction between two noncontiguous chromosome regions.
Alternatively, de novo formation of a centromere may simply begin with random imprinting or marking of a DNA sequence that favors the incorporation of some centromeric factors, such as the histone H3-like protein CENP-A. Once imprinted, the sequence can acquire additional proteins required for proper kinetochore assembly and propagate as a functional neocentromere regardless of the underlying nucleotide sequence composition. The initial state of the neocentromere may be imperfect for longterm evolutionary stability and may be subjected to further selection for improved kinetochore binding, for example, through repeated rounds of duplication of some crucial binding site, ultimately resulting in a new tandem array of repeats. Alternatively, as discussed above, the imperfect neocentromere may undergo an acquisition of a more preferred centromeric state through transposition of other centromeric or heterochromatic DNA.
Neocentromere Hotspots?
It is not known how a site is selected for neocentromere formation. A recent study on a series of inv dup (13q) chromosomes containing neocentromeres has identified specific “hotspots” for neocentromere activation at 13q32 and 13q21 (Warburton et al. 2000). It is possible that neocentromere sequences may share some sequence characteristics with one another and with α-satellite DNA, which has been shown to be a preferred substrate for centromere formation (Harrington et al. 1997; Ikeno et al. 1998). The development of a rapid method for identifying functionally critical neocentromere DNA, based on a combined chromatin immunoprecipitation and array analysis procedure described in Lo et al. (2001), should allow the in-depth investigation of this possibility through in silico analysis and comparison of a variety of cloned neocentromere DNA sequences.
Platero et al. (1999) have demonstrated that centromere competence can be an innate characteristic of DNA containing heterochromatic blocks, which may favor contacts for microtubule attachment and oscillation during mitosis. This study raises the possibility of chromosomal regions with heterochromatic properties being intrinsic sites for neocentromere activation. Henikoff et al. (2000) have also shown that the heterochromatic state facilitates the localization of CENP-A proteins in both Drosophila and humans. Furthermore, it has been reported that a dicentric Y chromosome is capable of forming a new constriction in the q-terminal heterochromatic region of the chromosome (Bukvic et al. 1996). Although most neocentromeres are localized at euchromatic regions, heterochromatin-associated proteins have been detected at neocentromeres, suggesting that neocentromeres, regardless of whether they originate from heterohromatic DNA, carry some properties of heterochromatin (Saffery et al. 2000). Thus, it appears that a genomic region with intrinsic heterochromatic properties, or the ability to acquire such properties, may be favored sites for neocentromerization.
Mechanisms for Accumulation of Repetitive DNA at Euchromatic Neocentromere
Recent molecular and cytological studies have identified the occurrence of duplication and transposition of region-specific low-copy repeat elements, or duplicons, at the pericentromeric regions of chromosomes (Eichler et al. 1997; Horvath et al. 2000a,b). Analysis of the molecular structure of a 160-kb DNA segment that separates α-satellite from non-α-satellite on human chromosome 16p11 reveals that this junction is composed of duplicated segments from Xq28 and 4q24 and that this DNA is found further amplified and spread to other pericentromeric regions, resulting in the formation of large blocks of sequences of high homology among nonhomologous chromosomes (Horvath et al. 2000b). Guy et al. (2000) and Jackson et al. (1999)have similarly identified distinct domains of duplicated sequences at the proximal regions flanking chromosome 10 centromere, indicating reorganization of human pericentromeric heterochromatin and evolution of a boundary between pericentromeric repeats and euchromatins. They suggest that pericentromeric duplication represents a transient intermediate of duplication of euchromatin into centromeric heterochromatin. Pericentromeric regions, therefore, may be preferred sites for recruitment of repeats and DNA transposition, leading to expansion of satellite blocks and the reconstruction of a more complex repeat structure that results in the accumulation of heterochromatic DNA at the repositioned centromeric sites described by Ventura et al. (2001).
Many Questions Remain Unanswered
The study of Ventura et al. (2001) and that of an earlier report from the same group (Montefalcone et al. 1999) represent the first attempts to understand the gain or loss of centromeres on a chromosome on an evolutionary scale. These occurrences result in the apparent repositioning of centromeres on chromosomes. Although the approach used by these investigators has clearly provided useful insights into various mechanistic possibilities, many unanswered questions remain. What triggers the need to reposition a centromere? How are neocentromeres activated and epigenetically inherited? Do the pericentromeric heterochromatin contribute to this activation? How has the heterochromatin evolved on a repositioned centromere? How are functional higher-order neocentromere structures produced by a diversity of underlying DNA sequences? What are the mechanisms for the loss of a pre-existing centromere on a chromosome carrying a repositioned centromere? There are, clearly, many experimental paths that can be followed in search of answers to these questions, including the use of different organisms ranging from single-cell eukaryotes (Saccharomyces cerevisiae and Schizosaccharomyces pombe) to the higher eukaryotes (such as Caenorhabditis elegans, Drosophila, Arabidopsis, and primates). The choice of primates as the subject of study by the Rocchi laboratory provides evolutionary data that fill a relevant and important gap to meet these challenging questions.
Acknowledgments
This work is supported by the National Health and Medical Research Council of Australia.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL choo{at}cryptic.rch.unimelb.edu.au; FAX 61-3-93481391.
-
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.183901.
- Cold Spring Harbor Laboratory Press