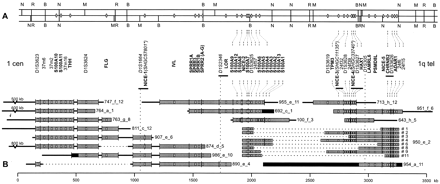
Integrated map of the EDC. Genomic and YAC restriction mapping results yielded the order of genes (in bold), STSs (in italics), and other loci within the EDC as shown. STS markers coinciding with the newly assigned genes were not tested and are marked with an asterisk. Precise localization of 37m2/37m16, of the two SPRR1 genes and ofS100A1/S100A13 was resolved previously (South et al. 1999). The order of loci underlined with a horizontal bar could not be resolved. (A) Genomic restriction map of the EDC. A continuous map spanning 3.5 Mb of region 1q21 with NotI (N),NruI (R), MluI (M), and BsiWI (B) restriction sites of the two haplotypes present in the H2LCL cell line (represented by the two lanes) is shown. White ovals indicate hybridization of the probe specified below to the corresponding restriction fragments that resulted from single and double restriction enzyme digests. Refined mapping results of S100A10, THH, FLG, IVL, SPRR3, SPRR1, SPRR2, LOR, S100A9, S100A8, and S100A6obtained by digestion with additional restriction enzymes (not shown) were adopted from the established map (Mischke et al. 1996). (B) Refined YAC contig of the EDC. Sixteen YACs from the 6-Mb contig covering the EDC (Marenholz et al. 1996) are shown with their addresses. Eight overlapping YACs of 950_e_2 that were used to resolve the order in the distal region are included. Sizes of YACs as determined previously by rotating-field gel electrophoresis (ROFE) are represented by the sum of the length of the gray and black boxes (detected fragments) and of the black horizontal bars (unidentified DNA sequences). White ovals represent hybridization of the respective fragment with the corresponding probe, derived from the locus above. SalI restriction sites are indicated by vertical lines. A polymorphic restriction site in NICE-1 was detected for 907_e_6 and 874_d_5. Gray boxes correspond to continuously covered human DNA as deduced from marker content and from identical fragments in several independent YACs. Broken horizontal lines indicate deletions and black boxes indicate other rearranged fragments identified by aberrant size and/or marker content. Restriction fragments of distinct size that were only detected in a single YAC are potential end fragments or could be rearranged and lack the second SalI site marker. Arrangement of detectedSalI restriction fragments with respect to the unidentified DNA sequences depended on YAC sizes and genomic mapping results.S100A3, S100A5, and S100A12 represented by gray ovals were mapped previously to YAC 100_f_3 (Schaefer et al. 1995; Wicki et al. 1996b). D1S1664, D1S2346, and D1S305 (vertical broken lines) were assigned to the corresponding YACs by PCR and therefore could not be positioned on single restriction fragments. The NICE-2, S100A7, and D1S3625 probes each hybridized to the same two SalI fragments, most likely indicating a duplication of sequences. Their order was resolved on the basis of signal intensities.











