Life with 25,000 Genes
Plants make the earth a good place for humans to live. They produce the oxygen we breathe, the food we eat, fuel for our cars and factories, fiber for the clothes we wear, wood for the houses we live in, and chemicals that keep us healthy or help cure us when we get sick. Plus, they are pleasant to look at, fun to grow, and intellectually interesting. Given all of this, it is surprising that they remain so poorly understood.
The completion of the Arabidopsis genome sequence will do much to change this ( Arabidopsis Genome Initiative 2000; Lin et al. 1999; Mayer et al. 1999; Salanoubat et al. 2000; Tabata et al. 2000;Theologis et al. 2000). As the first plant genome to be sequenced, this is rightly heralded as a landmark event. With the array of molecular genetic tools available for Arabidopsis and the impetus provided by the 2010 project, it will not be long before we know the physiological and developmental function of every gene in this species (Chory et al. 2000; Somerville and Dangl 2000). It is easy to take these events for granted in the era of genomic sequencing and reverse genetics. However, none of this would have happened without the major change in plant biology that has taken place in the last 15 years (Fink 1998). For example, in the late 1970's and early 1980's, students in Ian Sussex' plant biology laboratory completed Ph.D. theses on no less than 11 species, most of which were crop plants (my contribution was tobacco). At the time of Sussex's retirement in 1997, all but one of the students in his lab were studying Arabidopsis (I. Sussex, pers. comm.). The recent explosion of interest inArabidopsis—a weed of absolutely no economic importance—is unprecedented in the history of plant biology and provided this field with its first widely adopted model system. Arabidopsis is a small plant with a rapid life cycle and has been used in genetic studies for decades, but it was the discovery that Arabidopsishas a small genome with very little repeated DNA (Leutwiler et al. 1984; Pruitt and Meyerowitz 1986) that is primarily responsible for its recent popularity. The widespread adoption of Arabidopsis as an experimental system has resulted in rapid progress in many areas and has produced the international effort that led to the sequencing of theArabidopsis genome.
The Arabidopsis genome is 125 Mb and encodes ∼25,500 genes ( Arabidopsis Genome Initiative 2000). Arabidopsistherefore has significantly more genes than yeast (Goffeau et al. 1996), Caenorhabditis elegans ( C. elegans Sequencing Consortium 1998) or Drosophila (Adams et al. 2000). This is primarily because Arabidopsis genes often occur in more than one copy. Seventeen percent of the genes in Arabidopsis occur as tandem arrays of two or more closely related genes and ∼60% of the genome is segmentally duplicated, albeit in a highly rearranged fashion; the number of unique types of genes in Arabidopsis(12,000) is actually about equal to the number of gene types in worms (14,000) and flies (11, 000). The types of genes present inArabidopsis reinforce what has been learned from previous sequencing projects about the evolution of eukaryotes. Genes required for eukaryotic cell function such as components of the cytoskeleton, or essential processes such as DNA replication, repair and recombination, cell division, protein synthesis, and vesicle trafficking, are largely conserved between Arabidopsis and other eukaryotes. In contrast, genes involved in regulatory processes such as signal transduction and transcriptional regulation are quite different inArabidopsis, yeast, C. elegans, andDrosophila. For example, Arabidopsis has no genes similar to the components of major signaling pathways in animals, such as the Wingless/Wnt, Hedgehog, Notch/lin12, JAK/STAT, TGF-bβ/SMADS, and receptor tyrosine kinase/Ras pathways. Instead, signaling in Arabidopsis depends largely on receptor Ser/Thr kinases, of which there are 340, and a novel family of receptors related to bacterial two-component histidine kinases. Plants have also evolved a diverse array of transcription factors not found in animals. Arabidopsis has ∼1500 transcription factors, 1.3 times as many as in Drosophila and 1.7× as many as C. elegans or yeast (Riechmann et al. 2000). Forty-five percent of the families of transcription factors found in Arabidopsis are unique to plants. These differences should not be surprising, given that multicellularity originated independently in plants and animals, and plants have cell walls and animals do not (Meyerowitz 1999). The important lesson is that plants are as different from other organisms as they are the same, and are interesting for both reasons.
The fact that Arabidopsis has more genes than either C. elegans or Drosophila begs the question: Why does a structurally and behaviorally simple organism have more genes than organisms with a nervous system and cells that move? Although the answer to this question is unknown, there are some obvious possibilities. In contrast to animals, plants are autotrophs and can synthesize what they need to survive from air, light, water and a few mineral nutrients. In addition, they produce a huge variety of secondary compounds used for defense, disease resistance and a variety of other purposes; in fact, the number of secondary compounds produced by all plants is estimated to be as high as 100,000 ( Arabidopsis Genome Initiative 2000). Many of the enzymes that participate in these metabolic pathways are present in multiple copies in the genome. The extent to which this genetic diversity leads to functional diversity is unclear, but the potential for enormous biochemical diversity is certainly present in the Arabidopsisgenome. In short, animals are structurally more complex than plants, but plants probably do a lot more biochemistry. Another possible explanation for the difference in gene number between plants and animals is the remarkable ability of plants to deal with genetic imbalance. Variation in chromosome number resulting from either polyploidy or aneuploidy, as well as variation in gene dose due to segmental duplications and deficiencies, is tolerated much better by plants than by animals. Duplication and diversification of gene function may therefore be a more important source of novelty in plants than in animals. Of course, it is also possible that much of the genetic redundancy in Arabidopsis is functionally irrelevant.Arabidopsis may have more genes than either worms or flies because it can, not because it needs to.
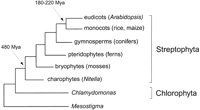
While it is interesting to compare Arabidopsis to species from which it diverged 1.6 billion years ago (Wang et al. 1999), the value of the Arabidopsis genome sequence lies primarily in what it reveals about plants. One surprise is the extent of segmental duplication ( Arabidopsis Genome Initiative 2000; Blanc et al. 2000; Vision et al. 2000). This observation confirms the results of interspecific comparisons of genome organization (Kowaleski et al. 1994; Paterson, 1996; Grant et al. 2000; Ku et al. 2000;) and suggests that the lineage leading to Arabidopsis underwent at least one genome duplication event, followed by extensive gene loss and rearrangement (Vision et al. 2000). Polyploidy is common in plants but was not predicted for a plant with five chromosomes and an unusually small genome. Estimates based on assumptions about the rate of amino acid substitution indicate that one large-scale duplication occurred ∼100 Myr ago, and suggest that other duplications may have occurred 140, 170, and 200 Myr ago (Vision et al. 2000). The latter duplications are old enough to have occurred prior to the divergence of major angiosperm lineages (including the split between monocots and dicots), and should therefore be shared by many different flowering plants (Figure 1). Extensive genome rearrangements can occur within a few generations in newly polyploid plants (Song et al. 1995), so it is unclear how useful information about genome structure will be for tracing evolutionary relationships. Nevertheless, the structure of the Arabidopsis genome will undoubtedly lead to a renewed appreciation of the importance of polyploidy in plant evolution (Soltis and Soltis 2000).
Knowing the identity of all 25,000 or so genes in Arabidopsisnow makes it possible to determine their function not only inArabidopsis but in other plant species as well. This is a large task because of the number of genes involved and the potential for functional redundancy but—at least in Arabidopsis—is technically not very difficult. Loss-of-function mutations can be readily identified by screening for transgene insertions by PCR (Krysan et al. 1996), and large populations of T-DNA transformed lines have been created specifically for this purpose. A variety of methods also exist for ectopically expressing genes in a regulated or unregulated fashion. Microarrays have already proven their value for determining global patterns of gene expression in Arabidopsis (Harmer et al. 2000), and will be an important tool for characterizing the effect of loss-of-function and gain-of-function mutations. The goal of determining the function of every gene in Arabidopsis by 2010 (Chory et al. 2000; Somerville and Dangl 2000) is therefore well within reach.
Information about the types of genes present in Arabidopsis is of widespread interest because gene function is often well conserved among flowering plants. Nevertheless, because many plants are highly polyploid and have experienced varying degrees of gene amplification, it is not necessarily a straightforward matter to identify orthologous genes in different species. It is also impossible to predict whether regulatory sequences have been conserved along with coding sequences. Analyses of the MADS box gene family illustrate this point. For example, Kramer and Irish (1998) showed that the evolution of two members of this family, PI and AP3, was accompanied by several duplication events and involved significant changes in gene expression patterns as well (Kramer and Irish 1999). Whether these changes are associated with changes in gene function is unknown. This phenomenon will be encountered over and over again as investigators try to move from Arabidopsis to other members of the plant kingdom and is a major challenge for future research.
The biggest obstacle to using Arabidopsis to identify genes in economically important plants is that most important crop plants in the world are monocots, and are therefore distantly related toArabidopsis. In order of harvested weight, the top 10 crops in 1999 were: sugarcane, maize, rice, wheat, potatoes, cassava, soybeans, sweet potatoes, and barley (see http://apps.fao.org/). Of these, sugarcane, maize, rice, wheat, and barley are monocots and members of the grass family. Ready access to the genomes of these species will come with the sequencing of the rice genome, expected to be completed in 10 years if not sooner (http://rgp.dna.affrc.go.jp/rgp/News/Newsletter.html). In the meantime, gene identification in these species is facilitated by the growing number of maize and rice ESTs and by other gene identification strategies that are being explored courtesy of a significant increase in federal funding for plant genomics. The value of sequencing the maize genome is controversial. In contrast to rice, maize has a large genome in which small islands of unique sequence are surrounded by oceans of repetitive DNA (Bennetzen et al. 1998); consequently, there is relatively little to be gained by sequencing more than only maize genes and their flanking 5′ and 3′ regions. On the other hand, the value of having a complete record of all the genes in the maize genome is undeniable, and many different ways to accomplish this goal are currently being explored (see http://plantgenome.sdsc.edu/).
After Arabidopsis, rice, and perhaps maize, what next? Although there is considerable interest in sequencing other crop species, it would be fundamentally more interesting (and, in the long run, perhaps more useful) to determine the genomic sequence of a unicellular ancestor of higher plants. This would provide a reference point for comparing the genomic organization of divergent plant lineages and would reveal the genetic basis for the morphological changes that accompanied the evolution of multicellularity (Graham et al. 2000). Flowering plants are members of the Streptophyta, a group that consists of all land plants and their closest algal relatives, the charophytes (Figure 1). Recent phylogenetic analyses based on ribosomal (Melkonian et al. 1995), actin (Bhattacharya et al. 1998), and chloroplast (Lemieux et al. 2000) gene sequences suggest that the unicellular biflagellate Mesostigma viridae is the most likely representative of the group from which streptophytes evolved. Consistent with its basal position, Mesostigma possesses a single actin gene (Arabidopsis has at least 10; McDowell et al. 1996) and has a larger complement of chloroplast genes than any other green alga or land plant. Efforts are already underway to sequence the genome of the unicellular chlorophyte,Chlamydomonas. These evolutionary arguments suggest that theMesostigma genome also has much to say about plant function and evolution, and should be given serious consideration.
With 25,000 mostly-uncharacterized genes, the Arabidopsisgenome will keep plant biologists busy for a long time. Given the traditional interest in plant diversity, it will be interesting to see whether this information will be used primarily to explore the biology of Arabidopsis, or as a starting point for forays into the far reaches of the plant kingdom. One thing is clear: With a host of interesting problems to study, an unrivalled set of molecular genetic tools, and now a sequenced genome, for plant biologists the fun has just begun.
Acknowledgments
I am grateful to Maja Bucan, Tony Cashmore, and members of my laboratory for helpful comments on this manuscript, and to Linda Graham and Claude Lemieux for information about Mesostigma viridae.
Footnotes
-
E-MAIL spoethig{at}sas.upenn.edu; FAX (215)898-8780.
-
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.
- Cold Spring Harbor Laboratory Press