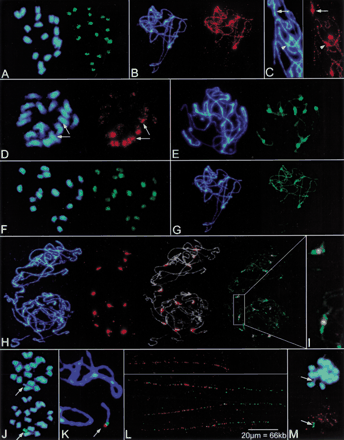
Physical mapping of centromere-associated, repetitive sequences onBeta chromosomes by fluorescent in situ hybridization (FISH). Blue fluorescence in panels shows the DNA stained with DAPI. (A) Hybridization of the Ty3-gypsy-like sequence pBv26 to mitotic metaphase chromosomes of B. vulgaris. Signals are largely confined to centromeric regions (green fluorescence). (B) Decondensed, paired chromosomes of B. procumbensat pachytene stage of meiosis. The Ty3-gypsy-like sequence pBp10 strongly hybridizes to the centromeric regions of five chromosome pairs (brightly stained with DAPI) and shows weak dispersion along chromosomes (red fluorescence). (C) Close-up image of the polymorphic pBp10 hybridization (red signal) to pachytene chromosomes of B. procumbens. The Ty3-gypsy-like sequence pBp10 shows confined amplification in the primary constriction (arrow) or clustering in the pericentromeric region with depletion in the constriction (arrowhead). (D) The satellite repeat pHC8 is detectable on three pairs of mitotic metaphase chromosomes of B. lomatogona (red fluorescence). Only one chromosome pair contains pHC8 arrays in the centromeric region (arrows) indicating polymorphisms of the centromeric heterochromatin between chromosomes in Betaspecies. (E) Pachytene chromosomes of B. vulgarisprobed with the satellite family pBV1 (green signals). The satellite forms large arrays within the DAPI-positive heterochromatin of all centromeres. (F) FISH with the oligonucleotide (GATA)4 to a mitotic metaphase plate of B. vulgarisshows that microsatellites are structural components amplified atBeta centromeres. (G) Rehybridization of B. procumbens pachytene chromosomes. After removal of the probe, chromosomes shown in (B) were hybridized with the satellite repeat pTS5 (strong green fluorescence). Note that the pTS5 satellite family colocalizes with DAPI-positive regions which also showed amplification of the Ty3-gypsy-like sequence pBp10. (H) Multicolor FISH of differently labeled satellite repeats to pachytene chromosomes of two cells of a heterozygous B. procumbens plant. The satellite family pTS5 (red signals) is amplified at most centromeres (middle left), the merged image (middle right) shows the morphology of decondensed pachytene chromosomes (gray) to indicate sites of hybridization. Examination with Filter 09 (right) shows that pTS4.1 satellite arrays (green signals) flank the prominent TS5 repeats (yellow). (I) Close examination of (H) showing the physical order of the pTS5 (yellow) and pTS4.1 (green) satellites at B. procumbens centromeres. (J) The chromosomal mutant PRO1 of B. vulgaris contains a B. procumbens chromosome fragment, which resembles a minichromosome. At mitotic metaphase, the PRO1 minichromosome is highly condensed, and both chromatides are visible (arrow). The minichromosome shows strong signals after FISH with the satellite repeat pTS5 (green fluorescence, arrow). (K) Close-up of FISH to pachytene chromosomes of PRO1. The arrow points to the minichromosome. Hybridization with the satellite pTS5 (red signal) and pTS4.1 (green signal) indicates that the centromeric region is close to the physical end of the minichromosome. (L) FISH to extended DNA fibers reveals the order of B. procumbens satellite repeats in the centromeric region of the PRO1 minichromosome. Stretches of red fluorescence indicate arrays of the satellite pTS5 (top panel). Note, that the pTS5 arrays are interrupted, detectable as gaps within the string of fluorescence signals. Double-target hybridization shows that the satellite pTS4.1 (green signals) forms an array adjacent to the longer pTS5 repeat block (red signals). Three examples are shown (lower panel). Yellow fluorescence indicates overlapping signals caused by interspersion of pTS4.1 repeats within gaps of the neighboring pTS5 array. (M) In the chromosomal mutant PAT2, the chromosome fragment from B. patellaris forms a minichromosome that is barely visible after DAPI staining (top, arrow) but is clearly detectable after FISH with the satellite repeat pTS5 (bottom, green fluorescence). Orange signals originate from simultaneous hybridization of the telomeric repeat (TTTAGGG).











