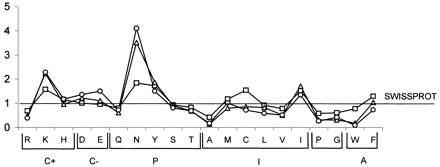
For Plasmodium falciparum chromosome-2–predicted proteins, ordinate values give the frequencies with which individual residues appear in complex regions (squares), nonrepetitive (triangles), and repetitive (circles) hydrophilic low-complexity segments, normalized with respect to frequencies averaged over the entire SWISS-PROT database (notes to release 38.0). Residues are grouped as follows: C+, positively charged; C−, negatively charged; P, uncharged polar; I, hydrophobic; A, aromatic nonpolar. Within groups, residues are ordered according to increasing hydrophobicity.











