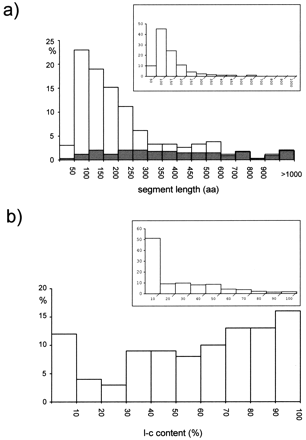
(a) Length distribution of the 415 low-complexity segments identified by the SEG program (window: 45; trigger: 3.4; extension: 3.75) in the complete set of 205 proteins predicted for Plasmodium falciparum chromosome 2. Superimposed in grey is the length distribution of the 77 internally repetitive low-complexity segments, always expressed as a percentage of the total number of SEG-identified segments. (Inset) Length distribution of the low-complexity segments identified by the SEG program (parameter setting as above) in the complete set of ORFs present on Saccharomyces cerevisiaechromosome II (Feldman et al. 1994). (b) Fractional distribution of the predicted proteins of P. falciparumchromosome 2 according to the percentage of the protein length occupied by low-complexity (l-c) segments, identified as above. (Insert) Idem for S. cerevisiae chromosome 2.











