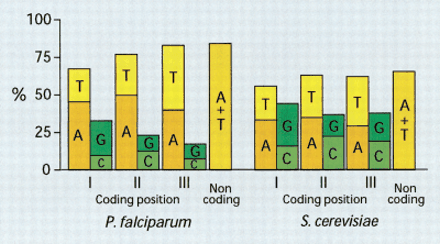
Nucleotide composition of coding and noncoding sequences is compared between chromosomes 2 of Plasmodium falciparum and Saccharomyces cerivisiae. It is evident that in both genomes the A +T content for each category reflects the general genomic bias. The detailed usage is, however, constrained by other selective pressures, particularly in codon positions I and II. In both organisms the A and T content of the third codon position is similar to noncoding regions. The frequency of T in the second codon position, which corresponds to hydrophobic amino acids, is ∼27% in both organisms but the frequency of A is much greater inPlasmodium (50%) than in yeast (35%). However, excluding low-complexity coding regions of Plasmodium gives a frequency of A (36%) similar to yeast, whereas low-complexity regions alone result in a frequency of 55%. Low-complexity regions inPlasmodium have a very different amino acid composition than yeast (Pizzi and Frontali 2001).











