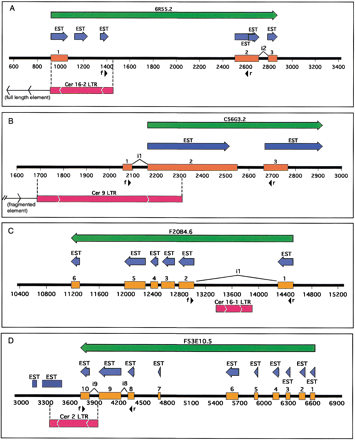
Cer element LTRs are part of some C. elegans genes. Green arrows represent Wormbase-predicted gene regions with corresponding identification. Blue arrows depict ESTs concordant to the predicted gene region. Orange boxes are predicted exon regions. Red boxes denote LTR position, and internal arrows indicate direction. The black line and numbers represent position along the genomic clone sequence (F20B4, C56G3, 6R55, F53E10). Black arrows indicate direction and location of forward (f) or reverse(r) PCR primers. For visual simplicity, only introns (i#) discussed in the text are displayed above and between exons. (A) An entire LTR from the 5′ end of a full-length Cer16-2 element is part of the 5′ end of a putative C. elegans gene (6R55.2) of unknown function. (B) The Cer9 LTR overlaps two exons of an aldo/keto reductase homolog in C. elegans (C56G3.2). The LTR is the 3′ end of a fragmented Cer9 element. (C) ACer16-1 solo LTR is part of intron 1 of a C. elegansgene (F20B4.6) in the glucosyltransferase family. (D) ACer2 solo LTR constitutes the 3′ end of a putative C. elegans gene (F53E10.5) of unknown function.











