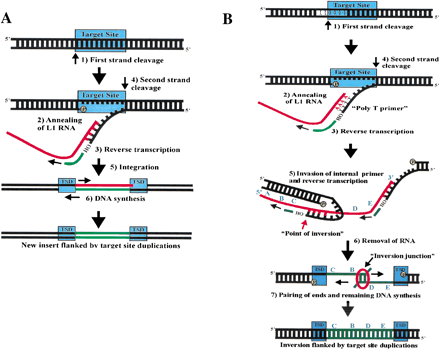
Target primed reverse transcription and twin priming. (A) This is a schematic of target primed reverse transcription (TPRT), based on in vitro studies of the R2 element from Bombyx mori (Luan et al. 1993). TPRT involves the following steps: (1) Cleavage of first DNA strand at the target site by the retrotransposon endonuclease (EN). (2) Annealing of retrotransposon RNA at the nick. (3) Reverse transcription from the free 3′-hydroxyl by the retrotransposon reverse transcriptase (RT). (4) Cleavage of second DNA strand. (5) Integration at the double-strand break. (6) Removal of RNA and completion of DNA synthesis. The TPRT process produces target site duplications (TSDs) at the flanks of the newly integrated retrotransposon. (B) Twin priming is a modification of the TPRT reaction with the following steps: (1) The L1 EN cleaves one strand of its DNA target site, producing the poly T primer. (2) The poly(A) tail of the L1 RNA anneals on the poly T primer. (3) L1 RT uses the L1 RNA as a template and the poly T primer to initiate reverse transcription. (4) The L1 EN cleaves the second DNA strand before reverse transcription has been completed, producing the internal primer. (5) The internal primer invades the L1 RNA and primes reverse transcription. (6) The RNA is removed from the RNA/cDNA structure. (7) The single-stranded cDNAs pair at a region of limited complementarity, and the remaining DNA synthesis is completed. The entire process results in an L1 inversion flanked by perfect target site duplications. The L1 RNA sequence is represented by 5′-A-B-C-D-E-3′. After the inversion, the insertion sequence is 5′-C-B-D-E-3′.











