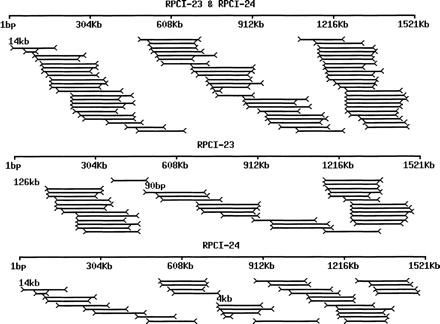
Effective clone coverage and gaps by paired ends of RPCI-23 and RPCI-24. The 1.5-Mb mouse chromosome 17 contig NT_002588‖Mm17_2723 was searched against a database consisting of 119,482 paired ends from RPCI-23 (7.8× clone coverage assuming an average insert size of 197 kb) and 72,433 paired ends from RPCI-24 (3.7× clone coverage assuming an average insert size of 155 kb). The searches were done in two steps as follows: (1) The contig sequence was searched against repeat-masked BESs by BLASTN; (2) the same sequence was researched against a mini-database consisting of unmasked paired ends from the clones identified by step one. Multiple hits were excluded and matches (1) with above, identity ≥ 95%; (2) match length ≥ 3/4 mBES length; (3) paired ends pointing toward each other; and (4) insert size ≤ 400 kb were selected. We have identified a total of 36 RPCI-23 pairs with a clone coverage of 4.6× and an average insert size of 191.6 kb (± 28.5 kb); and 33 RPCI-24 pairs with a clone coverage of 3.4× and an average insert size of 156 kb (± 39.7 kb). The paired-ends matches were plotted with the top for RPCI-23 and RPCI-24 (69 pairs with 7.9× coverage and one 14-kb gap), the middle for RPCI-23, and the bottom for RPCI-24. Bars represent the contig. Lines with arrows at both ends represent the matched pairs. Numbers represent the gaps.











