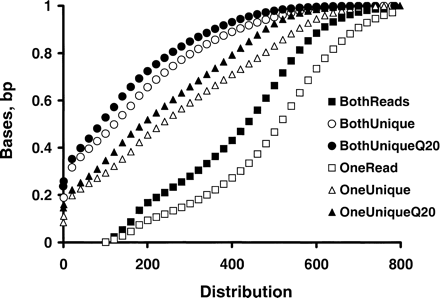
Length distributions of sequence reads, contiguous unique sequences, and contiguous unique Q20 bases of mBES paired ends. The unique bases and associated phred QVs were examined for 383,832 repeat masked mBESs (186 Mb) that consisted of paired ends from 119,483 RPCI-23 clones and 72,433 RPCI-24 clones. The numbers of read length, contiguous unique bases, and contiguous unique Q20 bases were counted for each mBES. The analyses indicated that all clones have their two BESs above 100 bp [(□) read-length distribution of the two BESs of each clone, OneRead]; [(▪) read length distribution of the shorter BES of each clone, BothReads]; 71% of the clones had at least one end with ≥ 100 bp contiguous unique sequences [(▵) OneUnique] and 54% had both ends with ≥ 100 bp contiguous unique sequences [(○) BothUnique]; 66% had at least one end with ≥ 100 bp contiguous unique Q20 bases [(▴) OneUniqueQ20] and 48% had both ends with ≥ 100 bp contiguous unique Q20 bases [(●) BothUniqueQ20].











