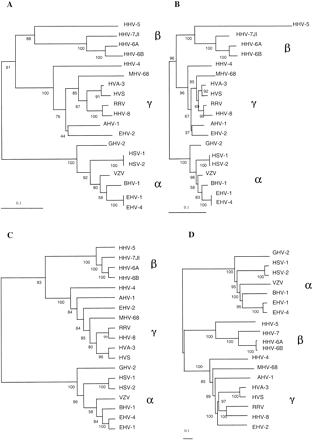
Phylogenetic trees based on protein family phylogenetic profiles (A,B,C) or on sequence comparison of herpesvirus-conserved domains (D). The initial data or alignments were bootstrapped 100 times. Neighbor-joining trees were constructed using the fraction of nonshared homologous families (A) or the fraction of dissimilar homologous families (B), as described in the text. Maximum parsimony cladogram built from the phylogenetic profiles (C). Neighbor-joining tree constructed using conserved regions in 26 herpesvirus open reading frames (D). α, Alphaherpesviruses; β, Betaherpesviruses; and γ, Gammaherpesviruses. The rhadinovirus subgroup of the Gammaherpesviruses consists of the viruses MHV-68, AHV-1, HVA-3, HVS, RRV, HHV-8, and EHV-2.











