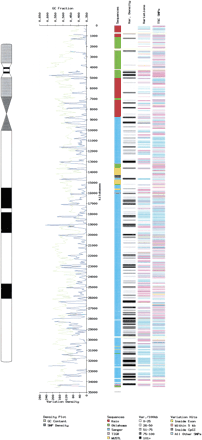
(following page) Distribution of polymorphisms on human chromosome 22. An ideogram of chromosome 22 with a schematic representation of the Giemsa banding pattern is shown at left. Next, the region containing the finished sequence is expanded to show the SNP map. The SNP density (the number of candidate variants in consecutive 100-kb regions) is plotted (blue line) superimposed on a plot of GC density (green). GC content is calculated as a percentage of the sequence using a sliding 100-kb window moved in 50-kb increments. The first column to the right of the graph represents sequences color-coded as per the collaborating institutions that contributed to the sequence pale yellow bars drawn horizontally across the map represent current gaps in the completed sequence. The next column to the right (Var. Density) shows a gray scale coding the number of potential variations recorded in the given 100-kb region. Such potential variations may be SNP, insertion, or deletion polymorphisms relative to the published reference sequence. The next column represents these variations as line annotations with color coding to represent the position of the variation relative to genomic features such as exons and CpG islands (a color key is on the diagram). The last column represents the corresponding map of recently published TSC (The SNP Consortium) SNPs reported by our group. The very high density of the overlap variations described in this paper (the Variations column) is not evident from the diagram because of the limits of the resolution. This diagram can also be viewed as a link from http://www.sanger.ac.uk/cgi-bin/humace/snp_search where a zoom facility allows regions to be enlarged to show the positions of individual SNPs relative to annotated exons and CpG islands.











