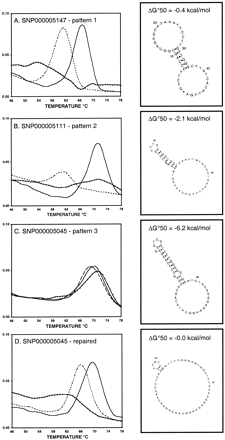
DASH results are shown for a major allele probe (solid line) and a minor allele probe (dashed line) used seperately to examine a major allele homozygous sample. Signals produced in the absence of any probe (representing target-sequence secondary structure) are also shown (diamond-marked line). All plots show the negative first derivative of the fluorescence versus temperature. The most stable target sequence secondary structures (predicted by mfold at 50°C) are shown to the right of each plot. HGBASE (http://hgbase.cgr.ki.se) SNP accession numbers are also given. (A) Pattern 1 (Ideal) assay. (B) Pattern 2 (Successful) assay. (C) Pattern 3 (Failed) assay. (D) An assay for the same SNP as inC, redesigned to minimize amplified target-sequence secondary structure as follows: original primers, 5′-GGCCTTCTCCCTGTAGATCCAC 3′ and biotin-5′-AGATACAGCACCAGCCTCAAGAC-3′; repaired primers, 5′-GGCCTTCTCCCTGTAGATCCAC-3′ and biotin-5′-ACAGAACAAGCCTCACGACGTCGG-3′.











