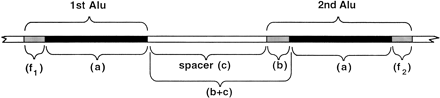
Diagram of an adjacent pair of Alus with aligned regions. TwoAlu sequences are presented that can be in inverted or direct-repeat orientation. The Alu sequences are identified by virtue of sharing homology with an Alu consensus sequence (see Methods). The alignment length (a) corresponds to a common (although nonidentical) region shared by both. External flanking sequences (f1) and (f2), if present, correspond to Alu regions of no (i.e., a truncation) or low similarity between the members of an Alupair. The spacer (c) consists of non-Alu sequence between the aligned regions and represents the minimal estimated distance between the two Alus. An internal flanking region is referred to as (b) so that the actual distance between aligned regions is the spacer (c) plus the internal flanking region (b) of an Alu. Therefore, the distance (b + c) is the actual distance between aligned regions of an adjacent pair of Alus.











