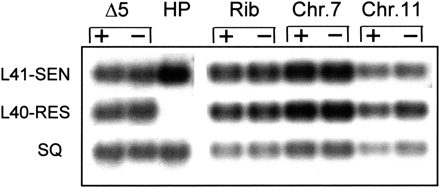
RT-PCR amplifications of dhfr/ts mRNAs from transfected clones harboring the dhfr/ts R gene in different chromosome locations—near the telomere in HPΔ5 parasites (Δ5); within C-type rRNA locus (Rib); and within the subtelomeric regions of chromosomes 7 and 11. Parasites were grown in the presence (+) or in the absence (−) of pyrimethamine selection. Hybridizations were performed with end-labeled oligonucleotides L41-SEN and L40-RES able to distinguish between the two versions of the dhfr/ts gene, as shown using the parental 8417HP as a control (HP), and probe SQ, which recognizes both transcripts, to normalize blotted DNA.











