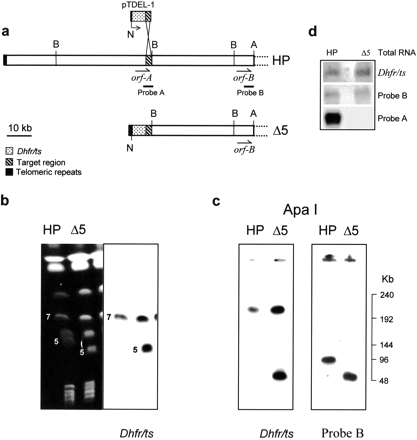
(a) Site-directed deletion strategy. A schematic map of the subtelomeric region of P. berghei chromosome 5 where the deletion is to occur is shown and the homology region with the transfecting plasmid is indicated. (A) ApaI; (N)NotI; (B) BamHI. (b) CHEF-separated chromosomes (left) of 8417HP and HPΔ5 (250 V, 55-sec pulse, 16 hr; 120 V, 270-sec pulse, 48 hr) blotted and hybridized withdhfr/ts probe (right). (c) 8417HP and HPΔ5 total DNA blocks digested with ApaI, electrophoresed by CHEF (250 V,12-sec pulse, 15 hr), blotted, and hybridized with the indicated probes. (d) Northern blot of total RNA from 8417HP (HP) and HPΔ5-clone 1 (Δ5) hybridized with dhfr/ts, probe A, and probe B. The latter has been used to normalize the blotted RNAs.











