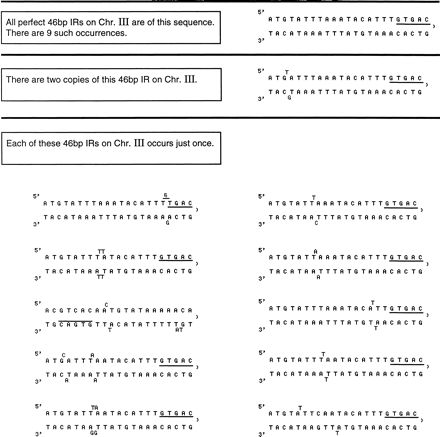
Figure 1.
The structures of all twenty-one 46 bp perfect or imperfect inverted repeats that contain the GTGAC motif on Caenorhabditis eleganschromosome III. Note that the drawings of the repeats are for the purpose of visualizing the symmetries and are not meant to imply that these occur in vivo or in vitro or that they have been analyzed thermodynamically for base pairing potential.











