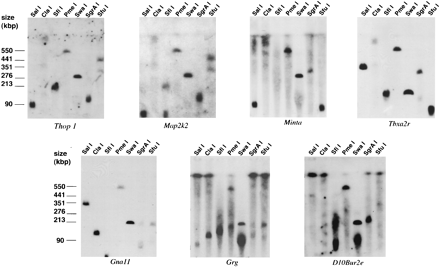
Pulsed-field gel electrophoresis (PFGE) mapping on mouse chromosome 10. DNA isolated in agarose plugs from mouse spleen was digested with a variety of restriction enzymes and separated in the 50–800 kb range on a BIORAD Chefmapper II using the conditions suggested by the autoalgorithm. Two filters were blotted from the same gel, and hybridized to radioactively labeled probes from mouse chromosome 10. Examples of overlapping hybridizing fragments are shown. Note that Thop1 and Map2k2 share many fragments, yet are >1000 kb apart on human 19p13.3, visualizing the inversion. Similarly, Grg and D10Bur2e share clearly many bands but are also ∼1000 kb apart in human.











