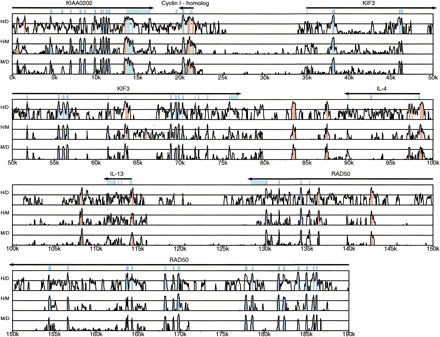
VISTA plot demonstrating peaks of similarity in the pairwise sequence alignments. Conserved sequences are shown relative to their positions in the human genome (horizontal axes), and their percent identities (50%–100%) are indicated on the vertical axes. The locations of coding exons (blue rectangles) and 5′- and 3′-untranslated regions (UTRs) (turquoise rectangles) are shown above the profile. Horizontal arrows indicate the direction of transcription for each gene. Peaks representing noncoding (red) and UTR (turquoise) sequences fitting the criteria for conserved elements as well as coding sequences (blue) meeting the percentage criteria over their entire length are indicated. The M/D alignment is mapped on the coordinates of the human genome sequence based on matching base pairs in the H/M alignment.











