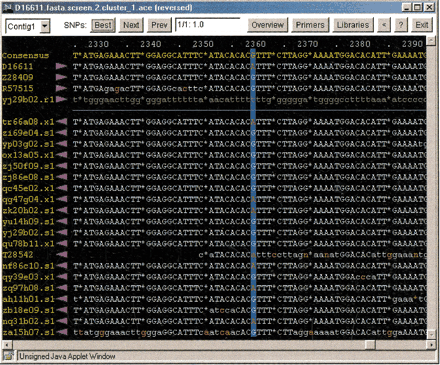
SNP viewer display of aligned sequences. In the window (black background), the consensus sequence of the assembly is displayed in yellow text. Gaps are indicated by asterisks. Coordinates of the assembly are above the consensus sequence in pale blue text. Individual sequences in the assembly are below in the consensus sequence. Sequence names are at the left in yellow text. Clicking on a sequence name launches a window containing a dbEST report for the sequence. The purple arrowhead to the right of a sequence name indicates the direction of the sequencing read. Sequences are to the right of the arrowheads. High quality basecalls are in uppercase pale blue text; low quality basecalls are in lowercase pale blue text; vector and low complexity sequences are in lowercase gray text. Basecalls differing from the consensus are in red. Clicking on a sequence launches a trace viewer. The candidate polymorphic site is indicated by dark blue vertical shading. In the panel at the top (white background), the Best, Next, and Prev buttons allow rapid navigation through the assembly to view candidate SNPs. The rank and probability score of the SNP are displayed in the small window at the center of the upper panel. The Overview button launches the assembly overview display (Fig. 4). The Primers button opens a link to the World Wide Web–based program Primer3 (Rozen and Skaletsky 1998), which can be used to design PCR primers for amplifying the region of DNA that contains the polymorphic site. The Libraries button opens a window listing the cDNA libraries that contain sequences defining the predicted SNP. Clicking the arrowhead button (<) reverses the contig.











