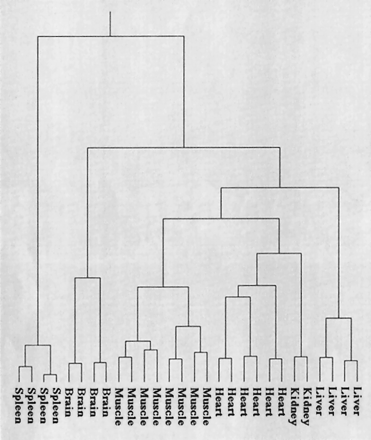
Clustering results for tissue-specific expression analysis. For generating the tissue-specific clustering tree or dendrogram, only slides showing an adequate hybridization quality were selected. The main criteria for judging the slide quality was the condition of the spotting blocks, meaning areas that were produced by a single spotting pin. The quality of a block was calculated by comparing the constantArabidopsis thaliana control clones to the mean background signal of the slides. Only slides with >90% of the blocks above the quality threshold were used for calculating the dendrogram, which corresponded to 14 out of 16 slides. Clustering was performed by the described hierarchical clustering method using a correlation-based distance measure for all clones on a slide.











