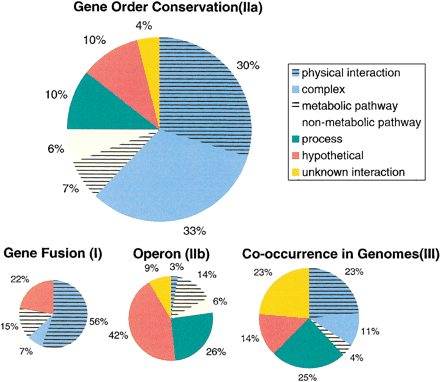
The types of functional interactions between M. genitaliumproteins for the different types of genomic context.
The surface areas of the circles are proportional to the number of genes for which the techniques apply. Classification was done by manual inspection, allowing detection of all possible described functional interactions between proteins. Subsequently, the functional interactions were divided along the following hierarchical classification:
1: direct physical interaction between the proteins 2: indirect physical interaction (i.e., the proteins are part of the same protein complex, but there is no evidence that they interact directly with each other) 3: the proteins are part of a single metabolic pathway 4: the proteins are part of a non-metabolic pathway, either regulatory or otherwise 5: the proteins take part in the same process 6: pairs of proteins of which at least one is hypothetical 7: proteins with known functions between which no functional interactions are known
Class 5 was only considered if the functional interactions between the proteins did not fall in classes 1–4. Types of functional interactions can be counted in two ways: per gene and per interaction. In general, the number of interactions is smaller than the number of genes, as two interacting genes only represent a single interaction. However, a single gene can have multiple genomic associations: In those cases they were normalized per gene. The results in the figure are based on a per gene count. The frequencies of the different classes of functional interactions did not alter significantly upon counting each interaction.











