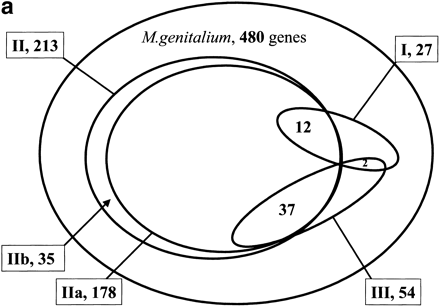
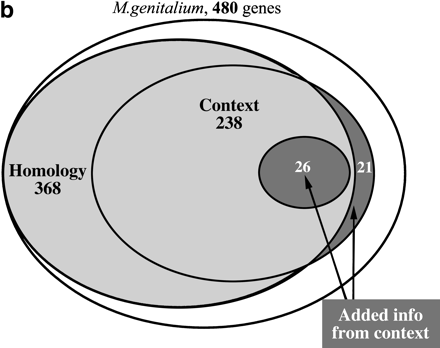
(a) Coverage of and overlap between various types of genomic context for M. genitalium genes. Type I is gene-fusion. Type II is the conservation local gene neighborhood, which is separated in type IIa (the conservation of gene order) and type IIb (the co-occurrence of genes within potential operons in absence of the conservation of gene order). Type III is the co-occurrence of genes in genomes. (b) Overlap between genes for which significant genomic context is available and genes for which functional features can be predicted by homology searches. For the latter, only genes that are homologous to genes with known molecular functions were included, which were determined by manual inspection. The dark gray areas in the figure are genes for which new functional features can be predicted by genomic context. They can be homologous to proteins with a known molecular function, in which case the context can indicate in which process this function plays a role (see text for specific examples). A complete list of genes for which new functional features could be predicted by genomic context and, if available, homology to proteins with known function, is available from http://dove.embl-heidelberg.de/MG/Context.











