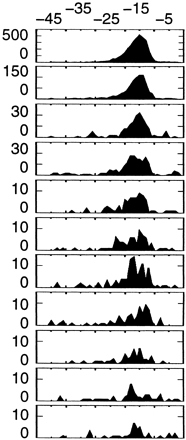
Table 2.
Most Significant Hexamers in 3′ Fragments: Clustered Hexamers
| Hexamer | Observed (expected) | % sites | P | Position average ± SD | Location | ||

|
3286 (317) | 58.2 | 0 | −16 ± 4.7 |
|
||
| 843 (112) | 14.9 | 0 | −17 ± 5.3 | ||||
| 156 (32) | 2.7 | 6 × 10−57 | −16 ± 5.9 | ||||
| 180 (53) | 3.2 | 4 × 10−45 | −18 ± 7.8 | ||||
| 76 (23) | 1.3 | 1 × 10−18 | −17 ± 5.9 | ||||
| 72 (21) | 1.3 | 2 × 10−18 | −18 ± 6.9 | ||||
| 96 (33) | 1.7 | 2 × 10−19 | −18 ± 6.9 | ||||
| 70 (16) | 1.2 | 5 × 10−23 | −18 ± 8.7 | ||||
| 43 (14) | 0.7 | 1 × 10−9 | −18 ± 6.3 | ||||
| 49 (11) | 0.8 | 5 × 10−17 | −18 ± 8.9 | ||||
| 36 (11) | 0.6 | 1 × 10−08 | −17 ± 8.1 | ||||
-
↵Expected occurrences based on a random distribution in a database of same nucleotide composition.
-
↵Probability of reaching the observed frequency by chance based on a cumulative binomial distribution. All sequences containing the most significant hexamer were removed from the database before the next most significant hexamer was sought.
-
↵Location of the last base of hexamer. Position 0 is the putative poly(A) site.











