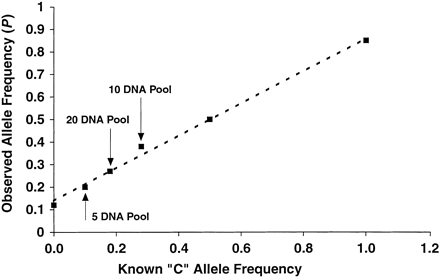
Allele frequency estimation for individual homozygotes, heterozygotes, and collections of multiple individuals at the SNP marker ANPex3.33. For the pooled samples, genomic DNA from a group of 5, 10, and 20 individuals (C-allele frequencies of 0.10, 0.28, and 0.18, respectively) was pooled in equal amounts and treated in the same way as the samples from single individuals. The observed allele fraction value P is plotted against the known C allele frequency, along with the best fit line as a guide to the eye. The line intercepts the Y-axis above the origin, and this systematic offset is the result of a small amount of cross-hybridization and misincorporation of the wrong base in the two-color SBE reaction. A correction can be applied to the data following the observation of pure genotypes to obtain a more accurate estimate of the absolute allele frequencies.











