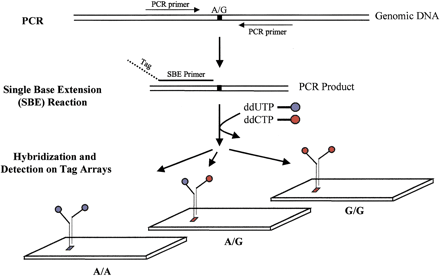
TAG–SBE genotyping assay. Marker-specific primers are designed for amplification of each SNP from genomic DNA (Wang et al. 1998); all SNPs with the same pair of variant bases (e.g., A/G SNPs) are pooled. The double-stranded PCR products serve as templates for the SBE reaction. Each SBE primer is chimeric with a 5' end complementary to a unique tag synthesized on the array and a 3' end complementary to the genomic sequence and terminating one base before a polymorphic SNP site. Thus, each SBE primer is uniquely associated with a specific tag (location) on the array. SBE primers corresponding to multiple markers are added to a single reaction tube and extended in the presence of pairs of ddNTPs labeled with different fluorophores; for example, an A/G bi-allelic marker is extended in the presence of biotin-labeled ddUTP and fluorescein-labeled ddCTP. The labeled multiplex SBE reaction products are pooled and hybridized to the tag array. Three hybridization patterns are shown, corresponding to three genotypes AA, AG, and GG.











