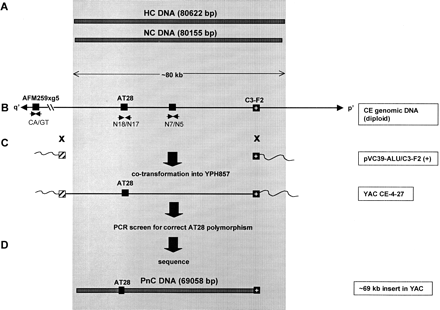
TAR cloning and sequencing of PnC DNA. The shaded area represents the region corresponding to the ∼80-kb 10q25.5 NC DNA (du Sart et al. 1997). (A) Sequenced regions of the HC DNA (derived from a CEPH library YAC clone) and NC DNA [derived from the mardel(10) neocentromere]. Total number of nucleotides sequenced is shown in brackets. (B) Structure of the HC/NC region and flanking DNA. Solid boxes represent STSs used in the identification and cloning of the DNA. AFM259xg5 is a (CA)n microsatellite located ∼150 kb (represented by the broken line) from the core region (Cancilla et al. 1998). AT28 (Barry et al. 1999) is a polymorphic VNTR used to identify the progenitor allele. C3-F2 is a 1.4-kb EcoRI fragment that served as the specific TAR “hook”(Cancilla et al. 1998). Small arrows indicate oligonucleotides used in PCR of the STSs. p′ and q′ refer to the short and long arms of mardel(10), respectively. (C) Radial TAR strategy using the vector pVC39-Alu/C3-F2(+) for the direct cloning of the progenitor DNA from the total genomic DNA of CE. The hatched box indicates the position of the Alu consensus sequence hook. Crosses denote the sites of recombination between the TAR vector pVC39-Alu/C3-F2( +) and CE genomic DNA at the C3-F2 andAlu hooks during cloning. The resulting circular YAC, CE-4–27, was shown by the AT28 polymorphism (see Fig. 3) to contain the PnC DNA from the progenitor chromosome 10. (D) The ∼69-kb sequenced portion of PnC DNA, represented by the bar.











