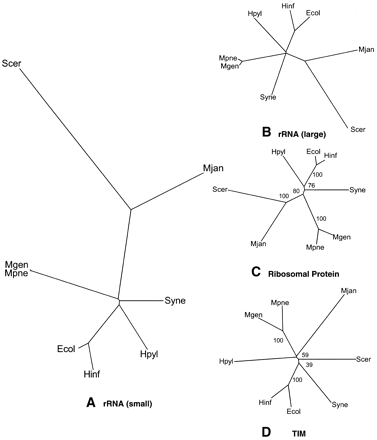
Representative single-gene trees. (A) The traditional small subunit ribosomal phylogenetic tree. This is a tree of eight completely sequenced representative organisms constructed with the SSU rRNA. Trees could be constructed using data from two different sources: the Ribosomal Database Project (RDP, http://www.cme.msu.edu/RDP, Maidak et al. 1999) and the rRNA WWW Server (http://www-rrna.uia.ac.be, Van de Peer et al. 1999). Although a tree can be abstracted from the RDP, the tree cannot contain both prokaryotes and eukaryotes. Instead, we took sequences from the RDP and the rRNA WWW server and aligned them with Clustal (Thompson et al. 1997). Phylip and PAUP were used to construct trees from the aligned sequences using distance and parsimony methods. There was little variation in the resulting trees displayed using TreeView (Page 1996), which was used to show all the trees used in this survey. The PAUP distance-based tree is shown. (B) The large subunit ribosomal tree. Another common method of building phylogenetic trees is the use of the large subunit rRNA (De Rijk et al. 1999). Because of the lack of large subunit rRNA information from the RDP, the sequences were downloaded from the rRNA WWW Server. The same method of tree construction was used as in A. The tree shown inB is the PAUP distance-based tree. Because of the large divergence of the species, the topology of the tree varied slightly when compared to the SSU ribosomal tree in A. The placement ofSynechocystis was slightly different, as it is placed closer to the eukaryote and Archeae in the large subunit tree. This was relatively less significant when considering the branch lengths of the tree in A . (C , D) Representative trees based on sequence similarity of orthologs. The sequences of proteins for the different organisms were obtained from the COGs web site (http://www.ncbi.nlm.nih.gov/COG, Tatusov et al. 1999). Clusters of orthologous groups were chosen that had one protein for each organism in the group. There were eight such COGs with representatives from four different classes. Distance-based trees and parsimony trees were both constructed for each of the orthologous groups. There was great variation in the resulting trees. The tree, which had the highest similarity to the traditional ribosomal tree, is shown in C. In fact, the distance-based tree based on the 30S ribosomal protein S3 (COG92, Class J) in C is exactly the same in topology to the traditional tree. This is not surprising because we expect a ribosomal protein tree to be similar to ribosomal rRNA trees because of their interaction and conservation. For the bootstrap values, all bootstrap replicates grouped E. coli with H. influenzaee,S. cerevisiae with M. jannaschii, and M. genitalium with M. pneumoniae. In all, the conserved topology coupled with high bootstrap values shows that phylogenetic trees with even a single protein can exhibit very high fidelity to the traditional ribosomal tree. Besides trees with high similarity to the traditional tree as in C, there were trees that varied significantly from the traditional ribosomal tree. Part Dshows a distance-based tree based on the metabolic enzyme triosephosphate isomerase (TIM). In general, there are a lot of differences between this tree and the traditional tree. M. jannaschii is grouped with M. genitalium and M. pneumoniae; M. jannaschii is not grouped with S. cerevisiae at all. The connectivity of S. cerevisiae andH. pylori is also different from the traditional tree. The low bootstrap values of 59% and 40% suggest that within the sequence there is great variation and the tree is generated with lower certainty. In general, there were a wide variety of trees produced using sequence similarity of orthologous proteins.











