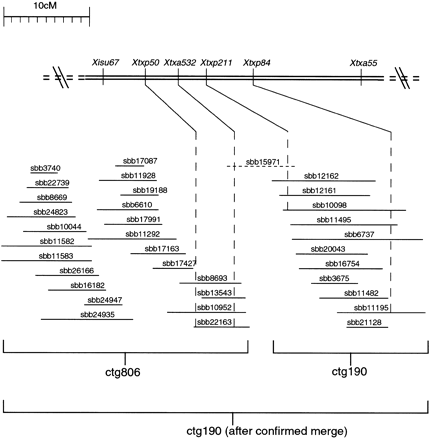
Integrated genetic and physical map of the region of sorghum LG B containing markers Xtxp50, Xtxa532, Xtxp211, and Xtxp84. The three SSR markers, Xtxp50, Xtxp211, and Xtxp84, were previously mapped to this region of LG B (G. Hart, pers. comm.), whereas the AFLP marker, Xtxa532, was mapped betweenXtxp50 and Xtxp211 in the present study. BACs linked to these four genetic markers by PCR-based screening of the BAC DNA pools are shown below the genetic map with the dashed linesextending from each marker through the respective, positive BAC clones. Four BAC clones in ctg806 were positive for Xtxa532 as well as the singleton BAC clone, sbb15971. Three BAC clones in ctg190 were positive for Xtxp211 as well as the singleton clone, sbb15971. Therefore, sbb15971 was used as a bridging clone to merge ctg190 and ctg806.











