Top Motifs Found by AlignACE
| (a) Most Specific Motifs Found by Aligning Sequence from Individual Organisms | |||||
| Known | Category | Organism | S site | MAP | Motif logo |
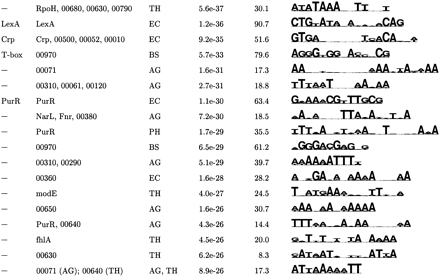
|
|||||
| (b) Most Specific Motifs Found by Aligning Sequence from Two or More Species | |||||
| Known | Category | Organism | S site | MAP | Motif Logo |
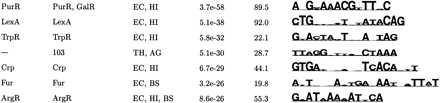
|
|||||
| (c) Top Palindromic Motifs Found in Sequence from Individual Organisms | |||||
| Known | Category | Organism | S site | MAP | Motif logo |
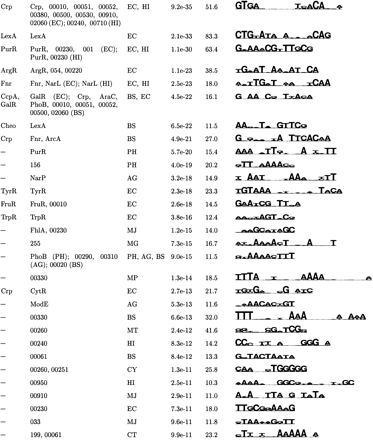
|
|||||
| (d) Top Palindromic Motifs Found by Aligning Sequence from Two or More Species | |||||
| Known | Category | Organism | S site | MAP | Motif logo |
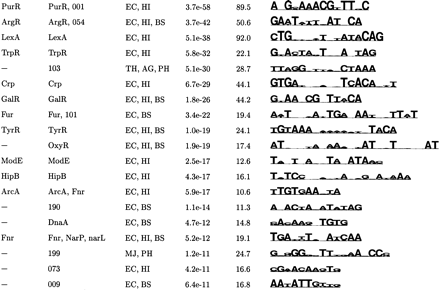
|
|||||
-
↵The top motifs were clustered according to their pairwise CompareACE scores, and the most specific member of each cluster is listed here (see Methods).
-
↵If this motif has been previously documenterd, its name is given.
-
↵The potentially coregulated group of genes in which this motif was found by AlignACE. For motifs found in categories derived from E. coli footprinted regulons, the name of the E. coli DNA-binding protein that regulates that regulon is listed. For motifs found in categories derived from conserved operons and KEGG functional group categories, either a 3-digit or 5-digit number is listed, respectively. These correspond to the following groups: KEGG functional group categories (84 total groups):
-
00010 Glycolysis/gluconeogenesis 00020 Citrate cycle (TCA) 00051 Fructose and mannose metabolism 00052 Galactose metabolism 00061 Fatty acid biosynthesis 00071 Fatty acid metabolism 00120 Bile acid biosynthesis 00220 Urea cycle and amino group metabolism 00230 Purine metabolism 00240 Pyrimidine metabolism 00251 Glutamate metabolism 00260 Glycine, serine, and threonine metabolism 00290 Valine, leucine, and isoleucine biosynthesis 00310 Lysine degradation 00330 Arginine and proline metabolism 00360 Phenylalanine metabolism 00380 Tryptophan metabolism 00500 Starch and sucrose metabolism 00530 Aminosugars metabolism 00630 Glyoxylate and dicarboxylate metabolism 00640 Propanoate metabolism 00650 Butanoate metabolism 00680 Methane metabolism 00710 Carbon fixation 00790 Folate biosynthesis 00910 Nitrogen metabolism 00950 Alkaloid biosynthesis I 00970 Aminoacyl-tRNA biosynthesis 02060 Phosphotransferase system (PTS)
-
Groups derived from conserved operons in WIT (343 total groups):
-
001 Purine biosynthesis 009 Nucleotide biosynthesis 033 Pyruvate synthase 054 Arginine biosynthesis 073 Fatty acid biosynthesis 101 Iron transport103 Ferrous ion transporter 156 HYP operon 190 Ribonucleoside diphosphate 199 Transcription 255 Heat shock
-
↵See Methods for abbreviations.
-
↵The Motif Logo (Schneider and Stevens, 1990). The height of the stack of letters is proportional to the information content, and the relative frequency of each base is given by its relative height. A sequence span containing the ten strongest positions of the logo is displayed.











