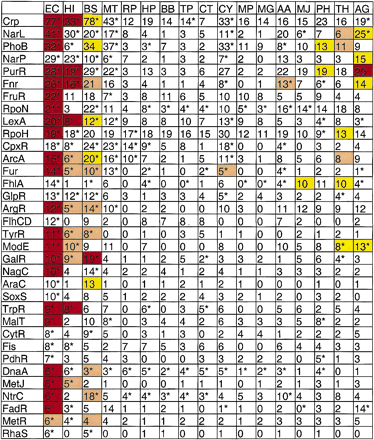
This figure summarizes the results in each organism from AlignACE runs on groups derived from E. coli footprinted regulons. The 34 known E. coli DNA-binding proteins that regulate at least five genes in E. coli are listed in the left column. The numbers indicate how many regulon members are present in that organism. (*) Ortholog to the E. coli DNA-binding protein was found. (Red or pink square) Motif similar to the known E. coli motif (CompareACE score >0.7) is conserved in this organism and was found by AlignACE with MAP score >5 and Ssite < 1e − 10. (Red square) Motif was found by aligning sequence from this organism only; (pink square) motif was found by aligning sequence from this organism together with sequence from E. coli (>30% of the aligned instances of the motif were from this organism). (Yellow square) New and significant motif found in sequence from this organism that is not similar to the E. coli motif. Yellow squares correspond to the motifs described in Tables 3a and 3c that were found in groups derived from E. coli regulons (i.e., these motifs score better than one of two cutoffs: (1) Ssite < 1e − 25, MAP score >10; (2) Ssite < 1e − 10, MAP score >10, palindromicity >0.7, % AT <80%. (White square) No new motif that scores better than these cutoffs, and the E. coli motif is not conserved in this organism.











