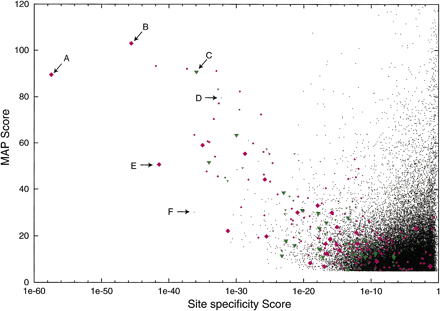
All of the motifs with MAP scores >5.0 (30,252 motifs) from the AlignACE runs in all 17 organisms are presented here (black points). The controls (E. coli motifs found in alignments of upstream regions of genes with >five footprints and which are known to constitute a regulon) are plotted with colored symbols: (green triangles) E. coli motifs found in alignments of upstream regions from genes making up the regulon in E. coli only; (magenta diamonds) conserved E. coli motifs found in alignment of sequence from E. coli together with sequence from B. subtilis, H. influenzae, or both. Several of the footprinted motifs are represented several times on this plot, as we perform two AlignACE runs on every group of genes, one for each of two different kinds of operon prediction. Also, a very strong motif can occasionally show up more than once in the same AlignACE run despite iterative masking in AlignACE. Therefore, for each control, we have represented the motif instance with the lowest value forSsite with a large colored symbol, and all other instances of the same motif with a small colored symbol. Several of the most specific motifs are labeled: (A) PurR conserved in E. coliand H. influenzae; (B) LexA conserved in E. coliand H. influenzae; (C) LexA in E. coli.; (D) T-box in B. subtilis; (E) ArgR conserved in E. coli., H. influenzae, and B. subtilis; (F) new motif found in the methane metabolism functional group in M. thermoautotrophicum. See text and Table 3 for details.











