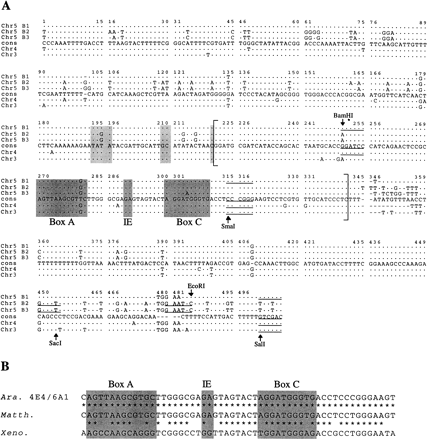
Alignment of the consensus sequences. (A) Alignment of the consensus sequences derived from the different 5S arrays and the consensus sequence of Campell et al. (1992). Restriction sites are indicated by arrows; conserved sequence elements upstream from the transcribed region (193–196, 209–210) and in the transcribed region BoxA, IE (Intermediate Element), and BoxC are indicated. The length of each consensus sequence is 501 bp, chromosome 3; 498 bp, chromosome 4; 502, 506, and 504 bp for chromosome 5 blocks 1, 2, and 3, respectively. The transcribed region is between brackets. (The SmaI site is not present in blocks 2 and 3, but contained in the primer sequence.) Dots (.) indicate identical residue; dashes (-) indicate a gap. (B) Homology between the internal promoter ofArabidopsis 5S rDNA (Ara 4E4/6A1) and the respective region of Matthiola incana (Matth) andXenopus 5S rDNA (Xeno).











