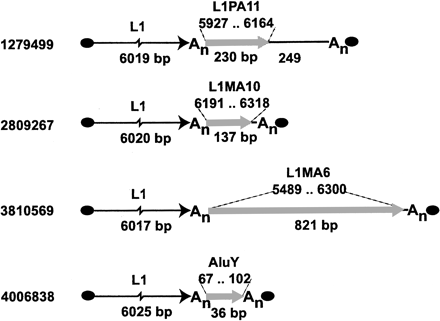
L1 integration into other transposons. Numbers at leftindicate gi numbers of GenBank records in which the L1 elements and their associated 3′-transduced segments are found (see Table 1 for exact coordinates). Repetitive elements recognized in the 3′-transduced segments by RepeatMasker are represented by filled gray arrows, with annotation shown at top (L1 or Alusubfamily name is followed by the coordinates projected onto the consensus sequence for that subfamily). Remarkably, in all four cases, the L1 that produced the observed new insertion depicted here must have inserted previously into another L1 or Alu in the same orientation. [A similar tendency has been noticed previously for newly inserted Alu elements (Jurka 1995)]. In two of the examples shown above, the transduced segment terminates at the end of the pre-existing L1. This suggests that once an L1 element inserts into such a region, subsequent transcription of the newly inserted element frequently reads through into the flanking DNA, and is then polyadenylated using a signal in the 3′ UTR of the pre-existing L1 element. By this means, L1 elements may acquire new, hybrid 3′ UTRs. This process likely contributes to the rapid evolution of L1 3′ UTR sequences (Smit 1996) and could explain why they are so rich in A residues, as well as provide a clue to the mechanism leading to the similarity of L1 and Alu 3′ UTR sequences (Boeke 1997).











