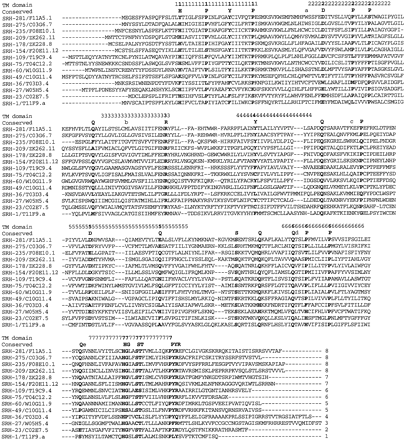
Alignment of the encoded amino acids of representative srhfamily chemoreceptors. One representative of each of the small basal subfamilies 1–4, two representatives each of subfamilies 5–7, and four representatives of subfamily 8 are shown, with the subfamily designations indicated at the end of the sequences. The seven transmembrane domains are indicated above the alignments following Fig.2. The alignments generally divide into blocks corresponding to these domains, with length variants between them, the exceptions being in the more variable TM4/5 region. The conserved amino acid positions that anchor the alignments are bold, and shown above the alignments, as are the inferred ancestral intron positions. Refined alignment of the highly variable amino and carboxyl termini is not shown.











