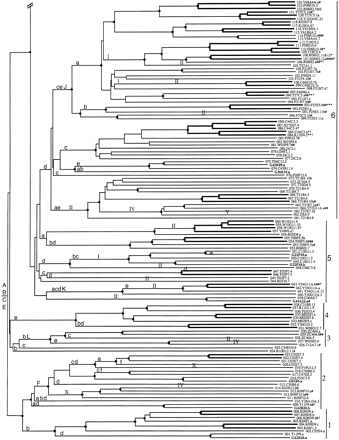

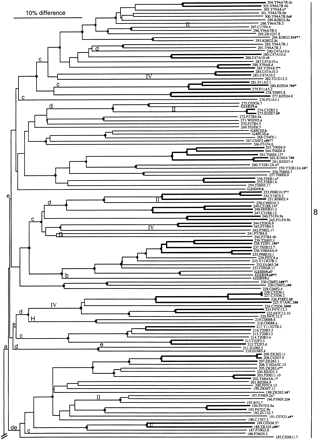
Phylogenetic tree relating members of the srh family of chemoreceptors. Subfamilies are indicated on the right by numbers. Bootstrap support >95% is indicated by a diamond on the relevant node, with a smaller circle indicating bootstrap support >75%. Inferred jumps of genes from chromosome V to another chromosome are indicated by roman numerals above the middle of the relevant branch. Lowercase letters above the base of the relevant branch indicate inferred intron loss, whereas uppercase letters indicate intron gain. Double-thickness lines connect genes that are inferred to have arisen by gene duplication since theelegans/briggsae species split. C. briggsae genes are indicated by bold type, all start with the letter G, and are not numbered. C. elegans genes are assigned gene numbers in asrh− series. Pseudogene status is indicated by symbols after each gene name. (#) Frameshift or large indel; (*) in-frame stop codon; (?) loss of start codon or questionable intron boundary.











