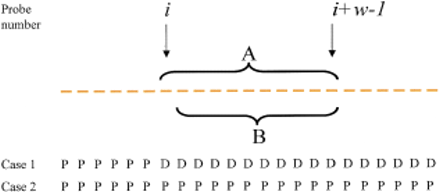
Enhancement of scores for hybridization by TSTEP is used to help define deletion regions. One iteration of the TSTEP calculation proceeds as follows: If the joint probability of (tandem) measurements A is less than the joint probability of (tandem) measurements B, and is also less than the probability of measurement i itself, then assign the probability of measurements A to position i. Note that A is the set of w measurements beginning at the ith measurement, where w is the window size. The score reassignment will tend to be different depending on whether probes query present (P) or deleted (D) regions. For example, in case 1, probei is the first probe to query in a deleted region (from left to right); it is quite likely that the probe will be reassigned a very low probability score. In case 2, where all probes query present DNA, probe i is likely either to not be re-assigned a score, or to be re-assigned a score which does not correspond to a very low probability.











