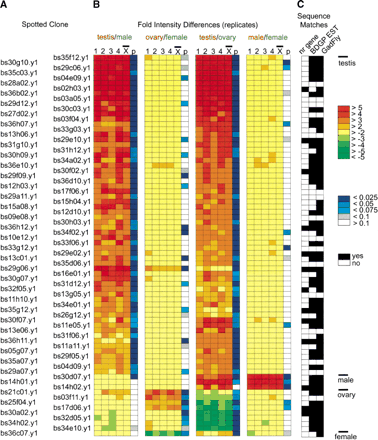
Summary of statistically significant microarray expression profiles (P <0.05, and threefold intensity difference). (A) Spotted cDNA clone name. (B) Microarray intensity differences from replicate comparisons (1, 2, 3, 4, and mean) of hybridizations with labeled cDNA from the indicated tissues. Tissues are color coded. For a given spotted cDNA, high relative hybridization with labeled “red” cDNA is indicated by red boxes, while high relative hybridization with labeled “green” cDNA is indicated by green boxes. Colorimetric scale is shown on the right. The P values for each spotted cDNA in each experimental series are indicated by blue color coded bars, with the scale shown on the right. The microarray cDNA clones are clustered into those showing testis (62), male (3), ovary (8), or female (1) preferential microarray expression profiles as indicated on the right. (C) A summary of sequence matches between ESTs from the respective microarray cDNAs and the indicated sequence databases. The black and white key is shown on the right (cutoffs are as follows: GenBank nr protein: BLASTX E-value <1E-20, and >90% sequence identity, BDGP ESTs and BDGP/CG predicted genes from the GadFly database: BLASTN E-value <1E-20).











