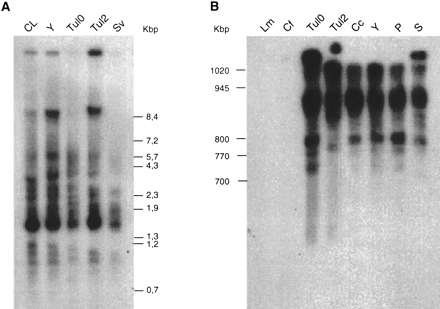
(A) Genomic DNA was prepared as described in Methods, digested with PstI and run in a 0.7% agarose gel at 3 V/cm. and transferred to a nylon membrane. (B) PFGE blots were prepared and processed as described (Henriksson et al. 1995). Chromosomal DNA markers were the CHEF DNA size markers, 0.2–2.2 Mbp (BioRad). Both nylon membranes (A,B) were hybridized with a radioactively labeled 300-bp fragment amplified by PCR using the oligonucleotides TcIRE-fwd and TcIRE-rev as shown in Figure 3 and described in Methods. Parasites, strains, and clones used in A or B are:Leishmania mexicana (Lm); Crithidia fasciculata (Cf);Trypanosoma cruzi strains and clones Tul0, Tul2, Corpus christi (Cc), Y, Perú (P), Sonya (S), CL-Brener (CL), and Sylvio (Sv).











